A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants
- PMID: 23803583
- PMCID: PMC3729767
- DOI: 10.1104/pp.113.217604
A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants
Abstract
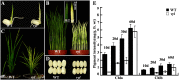
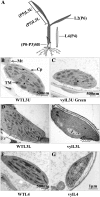
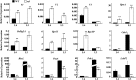
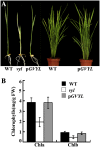
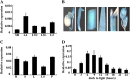
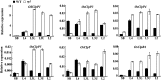
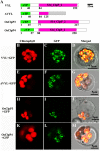
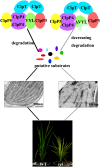
The plastidic caseinolytic protease (Clp) of higher plants is an evolutionarily conserved protein degradation apparatus composed of a proteolytic core complex (the P and R rings) and a set of accessory proteins (ClpT, ClpC, and ClpS). The role and molecular composition of Clps in higher plants has just begun to be unraveled, mostly from studies with the model dicotyledonous plant Arabidopsis (Arabidopsis thaliana). In this work, we isolated a virescent yellow leaf (vyl) mutant in rice (Oryza sativa), which produces chlorotic leaves throughout the entire growth period. The young chlorotic leaves turn green in later developmental stages, accompanied by alterations in chlorophyll accumulation, chloroplast ultrastructure, and the expression of chloroplast development- and photosynthesis-related genes. Positional cloning revealed that the VYL gene encodes a protein homologous to the Arabidopsis ClpP6 subunit and that it is targeted to the chloroplast. VYL expression is constitutive in most tissues examined but most abundant in leaf sections containing chloroplasts in early stages of development. The mutation in vyl causes premature termination of the predicted gene product and loss of the conserved catalytic triad (serine-histidine-aspartate) and the polypeptide-binding site of VYL. Using a tandem affinity purification approach and mass spectrometry analysis, we identified OsClpP4 as a VYL-associated protein in vivo. In addition, yeast two-hybrid assays demonstrated that VYL directly interacts with OsClpP3 and OsClpP4. Furthermore, we found that OsClpP3 directly interacts with OsClpT, that OsClpP4 directly interacts with OsClpP5 and OsClpT, and that both OsClpP4 and OsClpT can homodimerize. Together, our data provide new insights into the function, assembly, and regulation of Clps in higher plants.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

