Identifying the interaction between influenza and pneumococcal pneumonia using incidence data
- PMID: 23803706
- PMCID: PMC4178309
- DOI: 10.1126/scitranslmed.3005982
Identifying the interaction between influenza and pneumococcal pneumonia using incidence data
Abstract
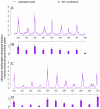
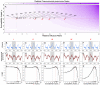
The association between influenza virus and the bacterium Streptococcus pneumoniae (pneumococcus) has been proposed as a polymicrobial system, whereby transmission and pathogenicity of one pathogen (the bacterium) are affected by interactions with the other (the virus). However, studies focusing on different scales of resolution have painted an inconsistent picture: Individual-scale animal experiments have unequivocally demonstrated an association, whereas epidemiological support in human populations is, at best, inconclusive. We integrate weekly incidence reports and a mechanistic transmission model within a likelihood-based inference framework to characterize the nature, timing, and magnitude of this interaction. We find support for a strong but short-lived interaction, with influenza infection increasing susceptibility to pneumococcal pneumonia ~100-fold. We infer modest population-level impacts arising from strong processes at the level of an individual, thereby resolving the dichotomy in seemingly inconsistent observations across scales. An accurate characterization of the influenza-pneumococcal interaction can form a basis for more effective clinical care and public health measures for pneumococcal pneumonia.
Figures


References
-
- Petney TN, Andrews RH. Multiparasite communities in animals and humans: Frequency, structure and pathogenic significance. Int. J. Parasitol. 1998;28:377–393. - PubMed
-
- Rohani P, Green C, Mantilla-Beniers N, Grenfell B. Ecological interference between fatal diseases. Nature. 2003;422:885–888. - PubMed
-
- Lello J, Boag B, Fenton A, Stevenson IR, Hudson PJ. Competition and mutualism among the gut helminths of a mammalian host. Nature. 2004;428:840–844. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

