Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila
- PMID: 23804096
- PMCID: PMC3693055
- DOI: 10.1523/JNEUROSCI.5419-12.2013
Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila
Abstract
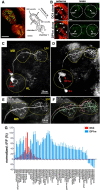
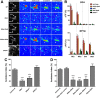
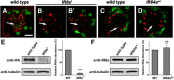
Drosophila olfactory sensory neurons express either odorant receptors or ionotropic glutamate receptors (IRs). The sensory neurons that express IR64a, a member of the IR family, send axonal projections to either the DC4 or DP1m glomeruli in the antennal lobe. DC4 neurons respond specifically to acids/protons, whereas DP1m neurons respond to a broad spectrum of odorants. The molecular composition of IR64a-containing receptor complexes in either DC4 or DP1m neurons is not known, however. Here, we immunoprecipitated the IR64a protein from lysates of fly antennal tissue and identified IR8a as a receptor subunit physically associated with IR64a by mass spectrometry. IR8a mutants and flies in which IR8a was knocked down by RNAi in IR64a+ neurons exhibited defects in acid-evoked physiological and behavioral responses. Furthermore, we found that the loss of IR8a caused a significant reduction in IR64a protein levels. When expressed in Xenopus oocytes, IR64a and IR8a formed a functional ion channel that allowed ligand-evoked cation currents. These findings provide direct evidence that IR8a is a subunit that forms a functional olfactory receptor with IR64a in vivo to mediate odor detection.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
