Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells
- PMID: 23804764
- PMCID: PMC3763554
- DOI: 10.1093/nar/gkt571
Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells
Abstract
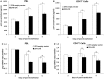
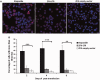
HIV-infected individuals currently cannot be completely cured because existing antiviral therapy regimens do not address HIV provirus DNA, flanked by long terminal repeats (LTRs), already integrated into host genome. Here, we present a possible alternative therapeutic approach to specifically and directly mediate deletion of the integrated full-length HIV provirus from infected and latently infected human T cell genomes by using specially designed zinc-finger nucleases (ZFNs) to target a sequence within the LTR that is well conserved across all clades. We designed and screened one pair of ZFN to target the highly conserved HIV-1 5'-LTR and 3'-LTR DNA sequences, named ZFN-LTR. We found that ZFN-LTR can specifically target and cleave the full-length HIV-1 proviral DNA in several infected and latently infected cell types and also HIV-1 infected human primary cells in vitro. We observed that the frequency of excision was 45.9% in infected human cell lines after treatment with ZFN-LTR, without significant host-cell genotoxicity. Taken together, our data demonstrate that a single ZFN-LTR pair can specifically and effectively cleave integrated full-length HIV-1 proviral DNA and mediate antiretroviral activity in infected and latently infected cells, suggesting that this strategy could offer a novel approach to eradicate the HIV-1 virus from the infected host in the future.
Figures



Similar articles
-
Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells.Mol Biol Rep. 2014 Sep;41(9):5819-27. doi: 10.1007/s11033-014-3456-3. Epub 2014 Jun 29. Mol Biol Rep. 2014. PMID: 24973878
-
HIV Provirus Stably Reproduces Parental Latent and Induced Transcription Phenotypes Regardless of the Chromosomal Integration Site.J Virol. 2016 May 12;90(11):5302-14. doi: 10.1128/JVI.02842-15. Print 2016 Jun 1. J Virol. 2016. PMID: 26984732 Free PMC article.
-
Zinc-Finger Nucleases Induced by HIV-1 Tat Excise HIV-1 from the Host Genome in Infected and Latently Infected Cells.Mol Ther Nucleic Acids. 2018 Sep 7;12:67-74. doi: 10.1016/j.omtn.2018.04.014. Epub 2018 May 3. Mol Ther Nucleic Acids. 2018. PMID: 30195798 Free PMC article.
-
New Approaches to Multi-Parametric HIV-1 Genetics Using Multiple Displacement Amplification: Determining the What, How, and Where of the HIV-1 Reservoir.Viruses. 2021 Dec 10;13(12):2475. doi: 10.3390/v13122475. Viruses. 2021. PMID: 34960744 Free PMC article. Review.
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
Cited by
-
Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells.Mol Biol Rep. 2014 Sep;41(9):5819-27. doi: 10.1007/s11033-014-3456-3. Epub 2014 Jun 29. Mol Biol Rep. 2014. PMID: 24973878
-
Nuclear landscape of HIV-1 infection and integration.Nat Rev Microbiol. 2017 Feb;15(2):69-82. doi: 10.1038/nrmicro.2016.162. Epub 2016 Dec 12. Nat Rev Microbiol. 2017. PMID: 27941817 Review.
-
Genome-Edited T Cell Therapies.Curr Stem Cell Rep. 2017;3(2):124-136. doi: 10.1007/s40778-017-0077-5. Epub 2017 Apr 18. Curr Stem Cell Rep. 2017. PMID: 28596938 Free PMC article. Review.
-
Lentivirus pre-packed with Cas9 protein for safer gene editing.Gene Ther. 2016 Jul;23(7):627-33. doi: 10.1038/gt.2016.27. Epub 2016 Apr 7. Gene Ther. 2016. PMID: 27052803
-
Genome editing of oncogenes with ZFNs and TALENs: caveats in nuclease design.Cancer Cell Int. 2018 Oct 22;18:169. doi: 10.1186/s12935-018-0666-0. eCollection 2018. Cancer Cell Int. 2018. PMID: 30386178 Free PMC article.
References
-
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. - PubMed
-
- Trono D, Van Lint C, Rouzioux C, Verdin E, Barré-Sinoussi F, Chun TW, Chomont N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

