Co-expression of RNA-protein complexes in Escherichia coli and applications to RNA biology
- PMID: 23804766
- PMCID: PMC3753655
- DOI: 10.1093/nar/gkt576
Co-expression of RNA-protein complexes in Escherichia coli and applications to RNA biology
Abstract
RNA has emerged as a major player in many cellular processes. Understanding these processes at the molecular level requires homogeneous RNA samples for structural, biochemical and pharmacological studies. We previously devised a generic approach that allows efficient in vivo expression of recombinant RNA in Escherichia coli. In this work, we have extended this method to RNA/protein co-expression. We have engineered several plasmids that allow overexpression of RNA-protein complexes in E. coli. We have investigated the potential of these tools in many applications, including the production of nuclease-sensitive RNAs encapsulated in viral protein pseudo-particles, the co-production of non-coding RNAs with chaperone proteins, the incorporation of a post-transcriptional RNA modification by co-production with the appropriate modifying enzyme and finally the production and purification of an RNA-His-tagged protein complex by nickel affinity chromatography. We show that this last application easily provides pure material for crystallographic studies. The new tools we report will pave the way to large-scale structural and molecular investigations of RNA function and interactions with proteins.
Figures

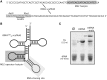
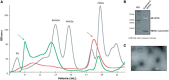
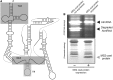


Similar articles
-
Coexpression and Copurification of RNA-Protein Complexes in Escherichia coli.Methods Mol Biol. 2021;2323:67-73. doi: 10.1007/978-1-0716-1499-0_6. Methods Mol Biol. 2021. PMID: 34086274
-
Expression and Purification of RNA-Protein Complexes in Escherichia coli.Methods Mol Biol. 2015;1316:25-31. doi: 10.1007/978-1-4939-2730-2_3. Methods Mol Biol. 2015. PMID: 25967050
-
A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold.Nat Protoc. 2009;4(6):947-59. doi: 10.1038/nprot.2009.67. Epub 2009 May 28. Nat Protoc. 2009. PMID: 19478810
-
Large scale expression and purification of recombinant RNA in Escherichia coli.Methods. 2011 Jun;54(2):267-73. doi: 10.1016/j.ymeth.2011.02.007. Epub 2011 Feb 12. Methods. 2011. PMID: 21320602 Review.
-
Biosynthesis and function of posttranscriptional modifications of transfer RNAs.Annu Rev Genet. 2012;46:69-95. doi: 10.1146/annurev-genet-110711-155641. Epub 2012 Aug 16. Annu Rev Genet. 2012. PMID: 22905870 Review.
Cited by
-
Advances in methods for native expression and purification of RNA for structural studies.Curr Opin Struct Biol. 2014 Jun;26:1-8. doi: 10.1016/j.sbi.2014.01.014. Epub 2014 Feb 28. Curr Opin Struct Biol. 2014. PMID: 24607442 Free PMC article. Review.
-
Intron-assisted, viroid-based production of insecticidal circular double-stranded RNA in Escherichia coli.RNA Biol. 2021 Nov;18(11):1846-1857. doi: 10.1080/15476286.2021.1872962. Epub 2021 Jan 20. RNA Biol. 2021. PMID: 33472518 Free PMC article.
-
Re-direction of carbon flux to key precursor malonyl-CoA via artificial small RNAs in photosynthetic Synechocystis sp. PCC 6803.Biotechnol Biofuels. 2018 Feb 5;11:26. doi: 10.1186/s13068-018-1032-0. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29441124 Free PMC article.
-
Co-expression and co-purification of archaeal and eukaryal box C/D RNPs.PLoS One. 2014 Jul 31;9(7):e103096. doi: 10.1371/journal.pone.0103096. eCollection 2014. PLoS One. 2014. PMID: 25078083 Free PMC article.
-
A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli.Sci Rep. 2018 Jan 30;8(1):1904. doi: 10.1038/s41598-018-20314-3. Sci Rep. 2018. PMID: 29382906 Free PMC article.
References
-
- Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

