Advancing genetic testing for deafness with genomic technology
- PMID: 23804846
- PMCID: PMC3887546
- DOI: 10.1136/jmedgenet-2013-101749
Advancing genetic testing for deafness with genomic technology
Abstract
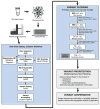
Background: Non-syndromic hearing loss (NSHL) is the most common sensory impairment in humans. Until recently its extreme genetic heterogeneity precluded comprehensive genetic testing. Using a platform that couples targeted genomic enrichment (TGE) and massively parallel sequencing (MPS) to sequence all exons of all genes implicated in NSHL, we tested 100 persons with presumed genetic NSHL and in so doing established sequencing requirements for maximum sensitivity and defined MPS quality score metrics that obviate Sanger validation of variants.
Methods: We examined DNA from 100 sequentially collected probands with presumed genetic NSHL without exclusions due to inheritance, previous genetic testing, or type of hearing loss. We performed TGE using post-capture multiplexing in variable pool sizes followed by Illumina sequencing. We developed a local Galaxy installation on a high performance computing cluster for bioinformatics analysis.
Results: To obtain maximum variant sensitivity with this platform 3.2-6.3 million total mapped sequencing reads per sample were required. Quality score analysis showed that Sanger validation was not required for 95% of variants. Our overall diagnostic rate was 42%, but this varied by clinical features from 0% for persons with asymmetric hearing loss to 56% for persons with bilateral autosomal recessive NSHL.
Conclusions: These findings will direct the use of TGE and MPS strategies for genetic diagnosis for NSHL. Our diagnostic rate highlights the need for further research on genetic deafness focused on novel gene identification and an improved understanding of the role of non-exonic mutations. The unsolved families we have identified provide a valuable resource to address these areas.
Keywords: deafness; hearing loss; massively parallel sequencing; sequence capture; targeted genomic enrichment.
Conflict of interest statement
SJ, HR, ACG, BN, SH, and EML are employees of Agilent Technologies Inc., which manufactures and offers for commercial sale targeted genomic enrichment kits like the one used in this work. All other others have no competing financial interests.
Figures

References
-
- Morton N. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630(1):16–31. - PubMed
-
- Schrauwen I, Sommen M, Corneveaux JJ, Reiman RA, Hackett NJ, Claes C, Claes K, Bitner-Glindzicz M, Couck P, Van Camp G, Huentelman MJ. A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am J Med Genet A. 2013;161A(1):145–152. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
