Are sucrose transporter expression profiles linked with patterns of biomass partitioning in Sorghum phenotypes?
- PMID: 23805151
- PMCID: PMC3693075
- DOI: 10.3389/fpls.2013.00223
Are sucrose transporter expression profiles linked with patterns of biomass partitioning in Sorghum phenotypes?
Abstract
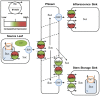
Sorghum bicolor is a genetically diverse C4 monocotyledonous species, encompassing varieties capable of producing high grain yields as well as sweet types which accumulate soluble sugars (predominantly sucrose) within their stems to high concentrations. Sucrose produced in leaves (sources) enters the phloem and is transported to regions of growth and storage (sinks). It is likely that sucrose transporter (SUT) proteins play pivotal roles in phloem loading and the delivery of sucrose to growth and storage sinks in all Sorghum ecotypes. Six SUTs are present in the published Sorghum genome, based on the BTx623 grain cultivar. Homologues of these SUTs were cloned and sequenced from the sweet cultivar Rio, and compared with the publically available genome information. SbSUT5 possessed nine amino acid sequence differences between the two varieties. Two of the remaining five SUTs exhibited single variations in their amino acid sequences (SbSUT1 and SbSUT2) whilst the rest shared identical sequences. Complementation of a mutant Saccharomyces yeast strain (SEY6210), unable to grow upon sucrose as the sole carbon source, demonstrated that the Sorghum SUTs were capable of transporting sucrose. SbSUT1, SbSUT4, and SbSUT6 were highly expressed in mature leaf tissues and hence may contribute to phloem loading. In contrast, SbSUT2 and SbSUT5 were expressed most strongly in sinks consistent with a possible role of facilitating sucrose import into stem storage pools and developing inflorescences.
Keywords: Sorghum; expression profiling; source–sink pathway; sucrose storage; sucrose transporters.
Figures








References
-
- Blum A., Sinmena B., Mayer J., Golan G., Shpiler L. (1994) Stem reserve mobilization supports wheat-grain filling under heat-stress. Aust. J. Plant Physiol. 21 771–781 10.1071/PP9940771 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

