Optimization of Multiple Pathogen Detection Using the TaqMan Array Card: Application for a Population-Based Study of Neonatal Infection
- PMID: 23805203
- PMCID: PMC3689704
- DOI: 10.1371/journal.pone.0066183
Optimization of Multiple Pathogen Detection Using the TaqMan Array Card: Application for a Population-Based Study of Neonatal Infection
Abstract
Identification of etiology remains a significant challenge in the diagnosis of infectious diseases, particularly in resource-poor settings. Viral, bacterial, and fungal pathogens, as well as parasites, play a role for many syndromes, and optimizing a single diagnostic system to detect a range of pathogens is challenging. The TaqMan Array Card (TAC) is a multiple-pathogen detection method that has previously been identified as a valuable technique for determining etiology of infections and holds promise for expanded use in clinical microbiology laboratories and surveillance studies. We selected TAC for use in the Aetiology of Neonatal Infection in South Asia (ANISA) study for identifying etiologies of severe disease in neonates in Bangladesh, India, and Pakistan. Here we report optimization of TAC to improve pathogen detection and overcome technical challenges associated with use of this technology in a large-scale surveillance study. Specifically, we increased the number of assay replicates, implemented a more robust RT-qPCR enzyme formulation, and adopted a more efficient method for extraction of total nucleic acid from blood specimens. We also report the development and analytical validation of ten new assays for use in the ANISA study. Based on these data, we revised the study-specific TACs for detection of 22 pathogens in NP/OP swabs and 12 pathogens in blood specimens as well as two control reactions (internal positive control and human nucleic acid control) for each specimen type. The cumulative improvements realized through these optimization studies will benefit ANISA and perhaps other studies utilizing multiple-pathogen detection approaches. These lessons may also contribute to the expansion of TAC technology to the clinical setting.
Conflict of interest statement
Figures

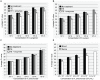
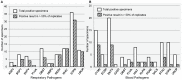

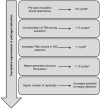
References
-
- Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, et al.. (2012) Etiology of Community-acquired Pneumonia among Hospitalized Adults in the United States: Preliminary Data from the CDC Etiology of Pneumonia in the Community (EPIC) Study, abstr 676.
-
- Jain S, Ampofo K, Arnold SR, Williams DJ, Anderson E, et al.. (2012) Etiology of Community-acquired Pneumonia among Hospitalized Children in the United States: Preliminary Data from the CDC Etiology of Pneumonia in the Community (EPIC) Study, abstr 168.
-
- Marrie TJ, Costain N, La SB, Patrick W, Forgie S, et al. (2012) The role of atypical pathogens in community-acquired pneumonia. Semin Respir Crit Care Med 33: 244–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

