Identification of Host Kinase Genes Required for Influenza Virus Replication and the Regulatory Role of MicroRNAs
- PMID: 23805279
- PMCID: PMC3689682
- DOI: 10.1371/journal.pone.0066796
Identification of Host Kinase Genes Required for Influenza Virus Replication and the Regulatory Role of MicroRNAs
Abstract
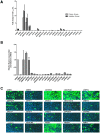
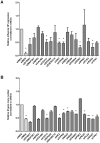
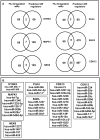
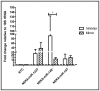
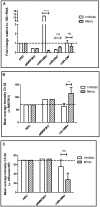
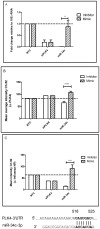
Human protein kinases (HPKs) have profound effects on cellular responses. To better understand the role of HPKs and the signaling networks that influence influenza virus replication, a small interfering RNA (siRNA) screen of 720 HPKs was performed. From the screen, 17 HPKs (NPR2, MAP3K1, DYRK3, EPHA6, TPK1, PDK2, EXOSC10, NEK8, PLK4, SGK3, NEK3, PANK4, ITPKB, CDC2L5 (CDK13), CALM2, PKN3, and HK2) were validated as essential for A/WSN/33 influenza virus replication, and 6 HPKs (CDK13, HK2, NEK8, PANK4, PLK4 and SGK3) were identified as vital for both A/WSN/33 and A/New Caledonia/20/99 influenza virus replication. These HPKs were found to affect multiple host pathways and regulated by miRNAs induced during infection. Using a panel of miRNA agonists and antagonists, miR-149* was found to regulate NEK8 expression, miR-548d-3p was found to regulate MAPK1 transcript expression, and miRs -1228 and -138 to regulate CDK13 expression. Up-regulation of miR-34c induced PLK4 transcript and protein expression and enhanced influenza virus replication, while miR-34c inhibition reduced viral replication. These findings identify HPKs important for influenza viral replication and show the miRNAs that govern their expression.
Conflict of interest statement
Figures
References
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, et al. (2004) Influenza-associated hospitalizations in the United States. JAMA 292: 1333–1340. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, et al. (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289: 179–186. - PubMed
-
- MMWR (2010) Estimates of Deaths Associated with Seasonal Influenza - United States, 1976–2007. Atlanta, GA: Centers for Disease Control and Prevention. 1057–1062 p. - PubMed
-
- Regoes RR, Bonhoeffer S (2006) Emergence of drug-resistant influenza virus: population dynamical considerations. Science 312: 389–391. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

