The duration of gastrin treatment affects global gene expression and molecular responses involved in ER stress and anti-apoptosis
- PMID: 23805861
- PMCID: PMC3698217
- DOI: 10.1186/1471-2164-14-429
The duration of gastrin treatment affects global gene expression and molecular responses involved in ER stress and anti-apoptosis
Abstract
Background: How cells decipher the duration of an external signal into different transcriptional outcomes is poorly understood. The hormone gastrin can promote a variety of cellular responses including proliferation, differentiation, migration and anti-apoptosis. While gastrin in normal concentrations has important physiological functions in the gastrointestine, prolonged high levels of gastrin (hypergastrinemia) is related to pathophysiological processes.
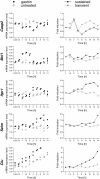
Results: We have used genome-wide microarray time series analysis and molecular studies to identify genes that are affected by the duration of gastrin treatment in adenocarcinoma cells. Among 403 genes differentially regulated in transiently (gastrin removed after 1 h) versus sustained (gastrin present for 14 h) treated cells, 259 genes upregulated by sustained gastrin treatment compared to untreated controls were expressed at lower levels in the transient mode. The difference was subtle for early genes like Junb and c-Fos, but substantial for delayed and late genes. Inhibition of protein synthesis by cycloheximide was used to distinguish between primary and secondary gastrin regulated genes. The majority of gastrin upregulated genes lower expressed in transiently treated cells were primary genes induced independently of de novo protein synthesis. This indicates that the duration effect of gastrin treatment is mainly mediated via post-translational signalling events, while a smaller fraction of the differentially expressed genes are regulated downstream of primary transcriptional events. Indeed, sustained gastrin treatment specifically induced prolonged ERK1/2 activation and elevated levels of the AP-1 subunit protein JUNB. Enrichment analyses of the differentially expressed genes suggested that endoplasmic reticulum (ER) stress and survival is affected by the duration of gastrin treatment. Sustained treatment exerted an anti-apoptotic effect on serum starvation-induced apoptosis via a PKC-dependent mechanism. In accordance with this, only sustained treatment induced anti-apoptotic genes like Clu, Selm and Mcl1, while the pro-apoptotic gene Casp2 was more highly expressed in transiently treated cells. Knockdown studies showed that JUNB is involved in sustained gastrin induced expression of the UPR/ER stress related genes Atf4, Herpud1 and Chac1.
Conclusion: The duration of gastrin treatment affects both intracellular signalling mechanisms and gene expression, and ERK1/2 and AP-1 seem to play a role in converting different durations of gastrin treatment into distinct cellular responses.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

