Inhibition of hepatitis B virus (HBV) gene expression and replication by HBx gene silencing in a hydrodynamic injection mouse model with a new clone of HBV genotype B
- PMID: 23805945
- PMCID: PMC3751867
- DOI: 10.1186/1743-422X-10-214
Inhibition of hepatitis B virus (HBV) gene expression and replication by HBx gene silencing in a hydrodynamic injection mouse model with a new clone of HBV genotype B
Abstract
Background: It has been suggested that different hepatitis B virus (HBV) genotypes may have distinct virological characteristics that correlate with clinical outcomes during antiviral therapy and the natural course of infection. Hydrodynamic injection (HI) of HBV in the mouse model is a useful tool for study of HBV replication in vivo. However, only HBV genotype A has been used for studies with HI.
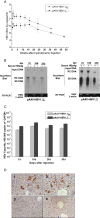
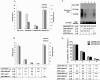
Methods: We constructed 3 replication-competent clones containing 1.1, 1.2 and 1.3 fold overlength of a HBV genotype B genome and tested them both in vitro and in vivo. Moreover, A HBV genotype B clone based on the pAAV-MCS vector was constructed with the 1.3 fold HBV genome, resulting in the plasmid pAAV-HBV1.3B and tested by HI in C57BL/6 mice. Application of siRNA against HBx gene was tested in HBV genotype B HI mouse model.
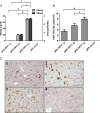
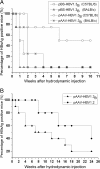
Results: The 1.3 fold HBV clone showed higher replication and gene expression than the 1.1 and 1.2 fold HBV clones. Compared with pAAV-HBV1.2 (genotype A), the mice HI with pAAV-HBV1.3B showed higher HBsAg and HBeAg expression as well as HBV DNA replication level but a higher clearance rate. Application of two plasmids pSB-HBxi285 and pSR-HBxi285 expressing a small/short interfering RNA (siRNA) to the HBx gene in HBV genotype B HI mouse model, leading to an inhibition of HBV gene expression and replication. However, HBV gene expression may resume in some mice despite an initial delay, suggesting that transient suppression of HBV replication by siRNA may be insufficient to prevent viral spread, particularly if the gene silencing is not highly effective.
Conclusions: Taken together, the HI mouse model with a HBV genotype B genome was successfully established and showed different characteristics in vivo compared with the genotype A genome. The effectiveness of gene silencing against HBx gene determines whether HBV replication may be sustainably inhibited by siRNA in vivo.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

