The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development
- PMID: 23806616
- PMCID: PMC3710456
- DOI: 10.1016/j.devcel.2013.05.022
The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development
Abstract
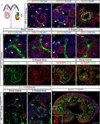
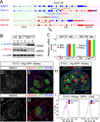
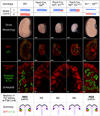
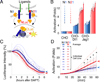
Notch2, but not Notch1, plays indispensable roles in kidney organogenesis, and Notch2 haploinsufficiency is associated with Alagille syndrome. We proposed that proximal nephron fates are regulated by a threshold that requires nearly all available free Notch intracellular domains (NICDs) but could not identify the mechanism that explains why Notch2 (N2) is more important than Notch1 (N1). By generating mice that swap their ICDs, we establish that the overall protein concentration, expression domain, or ICD amino acid composition does not account for the differential requirement of these receptors. Instead, we find that the N2 extracellular domain (NECD) increases Notch protein localization to the cell surface during kidney development and is cleaved more efficiently upon ligand binding. This context-specific asymmetry in NICD release efficiency is further enhanced by Fringe. Our results indicate that an elevated N1 surface level could compensate for the loss of N2 signal in specific cell contexts.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
The contribution of Notch1 to nephron segmentation in the developing kidney is revealed in a sensitized Notch2 background and can be augmented by reducing Mint dosage.Dev Biol. 2010 Jan 15;337(2):386-95. doi: 10.1016/j.ydbio.2009.11.017. Epub 2009 Nov 13. Dev Biol. 2010. PMID: 19914235 Free PMC article.
-
Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2.PLoS One. 2009 Aug 24;4(8):e6613. doi: 10.1371/journal.pone.0006613. PLoS One. 2009. PMID: 19701457 Free PMC article.
-
Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron.Development. 2007 Feb;134(4):801-11. doi: 10.1242/dev.02773. Epub 2007 Jan 17. Development. 2007. PMID: 17229764 Free PMC article.
-
Renal involvement and the role of Notch signalling in Alagille syndrome.Nat Rev Nephrol. 2013 Jul;9(7):409-18. doi: 10.1038/nrneph.2013.102. Epub 2013 Jun 11. Nat Rev Nephrol. 2013. PMID: 23752887 Review.
-
Alagille, Notch, and robustness: why duplicating systems does not ensure redundancy.Pediatr Nephrol. 2014 Apr;29(4):651-7. doi: 10.1007/s00467-013-2661-y. Epub 2013 Nov 24. Pediatr Nephrol. 2014. PMID: 24271660 Free PMC article. Review.
Cited by
-
Notch dimerization and gene dosage are important for normal heart development, intestinal stem cell maintenance, and splenic marginal zone B-cell homeostasis during mite infestation.PLoS Biol. 2020 Oct 5;18(10):e3000850. doi: 10.1371/journal.pbio.3000850. eCollection 2020 Oct. PLoS Biol. 2020. PMID: 33017398 Free PMC article.
-
Partitioning-Defective 1a/b Depletion Impairs Glomerular and Proximal Tubule Development.J Am Soc Nephrol. 2016 Dec;27(12):3725-3737. doi: 10.1681/ASN.2014111124. Epub 2016 May 16. J Am Soc Nephrol. 2016. PMID: 27185860 Free PMC article.
-
A perivascular niche for multipotent progenitors in the fetal testis.Nat Commun. 2018 Oct 30;9(1):4519. doi: 10.1038/s41467-018-06996-3. Nat Commun. 2018. PMID: 30375389 Free PMC article.
-
Notch signaling is required for the formation of mesangial cells from a stromal mesenchyme precursor during kidney development.Development. 2014 Jan;141(2):346-54. doi: 10.1242/dev.100271. Epub 2013 Dec 18. Development. 2014. PMID: 24353058 Free PMC article.
-
Presenilin gene function and Notch signaling feedback regulation in the developing mouse lens.Differentiation. 2018 Jul-Aug;102:40-52. doi: 10.1016/j.diff.2018.07.003. Epub 2018 Jul 25. Differentiation. 2018. PMID: 30059908 Free PMC article.
References
-
- Chen L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol. 2005;288:F939–F952. - PubMed
-
- Chu D, Zhang Z, Zhou Y, Wang W, Li Y, Zhang H, Dong G, Zhao Q, Ji G. Notch1 and Notch2 have opposite prognostic effects on patients with colorectal cancer. Ann Oncol. 2011;22:2440–2447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

