α-Amylase: an enzyme specificity found in various families of glycoside hydrolases
- PMID: 23807207
- PMCID: PMC11114072
- DOI: 10.1007/s00018-013-1388-z
α-Amylase: an enzyme specificity found in various families of glycoside hydrolases
Abstract
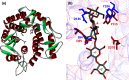
α-Amylase (EC 3.2.1.1) represents the best known amylolytic enzyme. It catalyzes the hydrolysis of α-1,4-glucosidic bonds in starch and related α-glucans. In general, the α-amylase is an enzyme with a broad substrate preference and product specificity. In the sequence-based classification system of all carbohydrate-active enzymes, it is one of the most frequently occurring glycoside hydrolases (GH). α-Amylase is the main representative of family GH13, but it is probably also present in the families GH57 and GH119, and possibly even in GH126. Family GH13, known generally as the main α-amylase family, forms clan GH-H together with families GH70 and GH77 that, however, contain no α-amylase. Within the family GH13, the α-amylase specificity is currently present in several subfamilies, such as GH13_1, 5, 6, 7, 15, 24, 27, 28, 36, 37, and, possibly in a few more that are not yet defined. The α-amylases classified in family GH13 employ a reaction mechanism giving retention of configuration, share 4-7 conserved sequence regions (CSRs) and catalytic machinery, and adopt the (β/α)8-barrel catalytic domain. Although the family GH57 α-amylases also employ the retaining reaction mechanism, they possess their own five CSRs and catalytic machinery, and adopt a (β/α)7-barrel fold. These family GH57 attributes are likely to be characteristic of α-amylases from the family GH119, too. With regard to family GH126, confirmation of the unambiguous presence of the α-amylase specificity may need more biochemical investigation because of an obvious, but unexpected, homology with inverting β-glucan-active hydrolases.
Figures




References
-
- Henrissat B, Davies GJ. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

