Specific gut commensal flora locally alters T cell tuning to endogenous ligands
- PMID: 23809163
- PMCID: PMC3748804
- DOI: 10.1016/j.immuni.2013.06.005
Specific gut commensal flora locally alters T cell tuning to endogenous ligands
Abstract
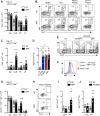
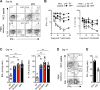
Differences in gut commensal flora can dramatically influence autoimmune responses, but the mechanisms behind this are still unclear. We report, in a Th1-cell-driven murine model of autoimmune arthritis, that specific gut commensals, such as segmented filamentous bacteria, have the ability to modulate the activation threshold of self-reactive T cells. In the local microenvironment of gut-associated lymphoid tissues, inflammatory cytokines elicited by the commensal flora dynamically enhanced the antigen responsiveness of T cells that were otherwise tuned down to a systemic self-antigen. Together with subtle differences in early lineage differentiation, this ultimately led to an enhanced recruitment of pathogenic Th1 cells and the development of a more severe form of autoimmune arthritis. These findings define a key role for the gut commensal flora in sustaining ongoing autoimmune responses through the local fine tuning of T-cell-receptor-proximal activation events in autoreactive T cells.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
-
- Allen A, Zheng Y, Gardner L, Safford M, Horton MR, Powell JD. The novel cyclophilin binding compound, sanglifehrin A, disassociates G1 cell cycle arrest from tolerance induction. J Immunol. 2004;172:4797–4803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

