The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy
- PMID: 23810512
- PMCID: PMC3720741
- DOI: 10.1016/j.devcel.2013.05.024
The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy
Abstract
As the cellular power plant, mitochondria play a significant role in homeostasis. To maintain the proper quality and quantity of mitochondria requires both mitochondrial degradation and division. A selective type of autophagy, mitophagy, drives the degradation of excess or damaged mitochondria, whereas division is controlled by a specific fission complex; however, the relationship between these two processes, especially the role of mitochondrial fission during mitophagy, remains unclear. In this study, we report that mitochondrial fission is important for the progression of mitophagy. When mitophagy is induced, the fission complex is recruited to the degrading mitochondria through an interaction between Atg11 and Dnm1; interfering with this interaction severely blocks mitophagy. These data establish a paradigm for selective organelle degradation.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



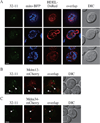


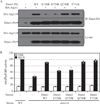
Comment in
-
Mitochondrial fission facilitates mitophagy in Saccharomyces cerevisiae.Autophagy. 2013 Nov 1;9(11):1900-1. doi: 10.4161/auto.25804. Epub 2013 Sep 4. Autophagy. 2013. PMID: 24025250 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

