The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament
- PMID: 23810717
- PMCID: PMC3991727
- DOI: 10.1016/j.dnarep.2013.05.005
The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament
Abstract
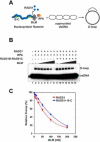
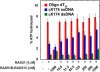
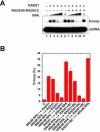
There are six human RAD51 related proteins (HsRAD51 paralogs), HsRAD51B, HsRAD51C, HsRAD51D, HsXRCC2, HsXRCC3 and HsDMC1, that appear to enhance HsRAD51 mediated homologous recombinational (HR) repair of DNA double strand breaks (DSBs). Here we model the structures of HsRAD51, HsRAD51B and HsRAD51C and show similar domain orientations within a hypothetical nucleoprotein filament (NPF). We then demonstrate that HsRAD51B-HsRAD51C heterodimer forms stable complex on ssDNA and partially stabilizes the HsRAD51 NPF against the anti-recombinogenic activity of BLM. Moreover, HsRAD51B-HsRAD51C stimulates HsRAD51 mediated D-loop formation in the presence of RPA. However, HsRAD51B-HsRAD51C does not facilitate HsRAD51 nucleation on a RPA coated ssDNA. These results suggest that the HsRAD51B-HsRAD51C complex plays a role in stabilizing the HsRAD51 NPF during the presynaptic phase of HR, which appears downstream of BRCA2-mediated HsRAD51 NPF formation.
Keywords: ATPase, Bloom's syndrome; BLM; Homologous recombination; RAD51 paralogs.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures




References
-
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. - PubMed
-
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. - PubMed
-
- Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

