Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens
- PMID: 23810758
- PMCID: PMC4912568
- DOI: 10.1016/j.jmoldx.2013.05.004
Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens
Abstract
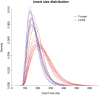
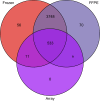
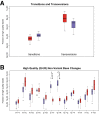
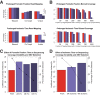
Next-generation sequencing (NGS) has emerged as a powerful technique for the detection of genetic variants in the clinical laboratory. NGS can be performed using DNA from FFPE tissue, but it is unknown whether such specimens are truly equivalent to unfixed tissue for NGS applications. To address this question, we performed hybridization-capture enrichment and multiplexed Illumina NGS for 27 cancer-related genes using DNA from 16 paired fresh-frozen and routine FFPE lung adenocarcinoma specimens and conducted extensive comparisons between the sequence data from each sample type. This analysis revealed small but detectable differences between FFPE and frozen samples. Compared with frozen samples, NGS data from FFPE samples had smaller library insert sizes, greater coverage variability, and an increase in C to T transitions that was most pronounced at CpG dinucleotides, suggesting interplay between DNA methylation and formalin-induced changes; however, the error rate, library complexity, enrichment performance, and coverage statistics were not significantly different. Comparison of base calls between paired samples demonstrated concordances of >99.99%, with 96.8% agreement in the single-nucleotide variants detected and >98% accuracy of NGS data when compared with genotypes from an orthogonal single-nucleotide polymorphism array platform. This study demonstrates that routine processing of FFPE samples has a detectable but negligible effect on NGS data and that these samples can be a reliable substrate for clinical NGS testing.
Copyright © 2013 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25:795–804. - PubMed
-
- Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A.S., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L., Novetsky A.P., Jarvik G.P., Olopade O.I., Goodfellow P.J., King M.C., Tait J.F., Walsh T. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. - PMC - PubMed
-
- Walter M.J., Shen D., Ding L., Shao J., Koboldt D.C., Chen K., Larson D.E., McLellan M.D., Dooling D., Abbott R., Fulton R., Magrini V., Schmidt H., Kalicki-Veizer J., O'Laughlin M., Fan X., Grillot M., Witowski S., Heath S., Frater J.L., Eades W., Tomasson M., Westervelt P., DiPersio J.F., Link D.C., Mardis E.R., Ley T.J., Wilson R.K., Graubert T.A. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

