The Role of Aromatic-Aromatic Interactions in Strand-Strand Stabilization of β-Sheets
- PMID: 23810905
- PMCID: PMC3778025
- DOI: 10.1016/j.jmb.2013.06.030
The Role of Aromatic-Aromatic Interactions in Strand-Strand Stabilization of β-Sheets
Abstract
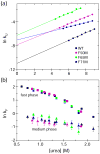
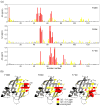
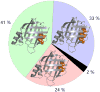
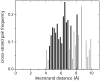
Aromatic-aromatic interactions have long been believed to play key roles in protein structure, folding, and binding functions. However, we still lack full understanding of the contributions of aromatic-aromatic interactions to protein stability and the timing of their formation during folding. Here, using an aromatic ladder in the β-barrel protein, cellular retinoic acid-binding protein 1 (CRABP1), as a case study, we find that aromatic π stacking plays a greater role in the Phe65-Phe71 cross-strand pair, while in another pair, Phe50-Phe65, hydrophobic interactions are dominant. The Phe65-Phe71 pair spans β-strands 4 and 5 in the β-barrel, which lack interstrand hydrogen bonding, and we speculate that it compensates energetically for the absence of strand-strand backbone interactions. Using perturbation analysis, we find that both aromatic-aromatic pairs form after the transition state for folding of CRABP1, thus playing a role in the final stabilization of the β-sheet rather than in its nucleation as had been earlier proposed. The aromatic interaction between strands 4 and 5 in CRABP1 is highly conserved in the intracellular lipid-binding protein (iLBP) family, and several lines of evidence combine to support a model wherein it acts to maintain barrel structure while allowing the dynamic opening that is necessary for ligand entry. Lastly, we carried out a bioinformatics analysis and found 51 examples of aromatic-aromatic interactions across non-hydrogen-bonded β-strands outside the iLBPs, arguing for the generality of the role played by this structural motif.
Keywords: ASA; COM; CRABP1; FABP; H-bond; HSQC; IFABP; ILBP; MD; MSA; PDB; Protein Data Bank; TS; WT; accessible surface area; cellular retinoic acid-binding protein 1; center of mass; dynamics; fatty acid-binding protein; folding; heteronuclear single quantum coherence; hydrogen bond; iLBP; ileal lipid-binding protein; intestinal fatty acid-binding protein; intracellular lipid-binding protein; molecular dynamics; multiple sequence alignment; transition state; wild type; β-barrel; π stacking.
© 2013 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Sequence and structural analysis of cellular retinoic acid-binding proteins reveals a network of conserved hydrophobic interactions.Proteins. 2004 Feb 1;54(2):179-94. doi: 10.1002/prot.10520. Proteins. 2004. PMID: 14696180
-
Local and non-local topological information in the denatured state ensemble of a β-barrel protein.Protein Sci. 2018 Dec;27(12):2062-2072. doi: 10.1002/pro.3516. Epub 2018 Oct 16. Protein Sci. 2018. PMID: 30252171 Free PMC article.
-
Evolutionary coupling of structural and functional sequence information in the intracellular lipid-binding protein family.Proteins. 2006 May 1;63(2):373-84. doi: 10.1002/prot.20860. Proteins. 2006. PMID: 16477649
-
Intervening in the β-barrel structure of lipid binding proteins: consequences on folding, ligand-binding and aggregation propensity.Prostaglandins Leukot Essent Fatty Acids. 2015 Feb;93:37-43. doi: 10.1016/j.plefa.2014.08.001. Epub 2014 Aug 11. Prostaglandins Leukot Essent Fatty Acids. 2015. PMID: 25242388 Review.
-
Geometry of nonbonded interactions involving planar groups in proteins.Prog Biophys Mol Biol. 2007 Sep-Nov;95(1-3):83-137. doi: 10.1016/j.pbiomolbio.2007.03.016. Epub 2007 Jun 5. Prog Biophys Mol Biol. 2007. PMID: 17629549 Review.
Cited by
-
The conserved aromatic residue W122 is a determinant of potyviral coat protein stability, replication, and cell-to-cell movement in plants.Mol Plant Pathol. 2021 Feb;22(2):189-203. doi: 10.1111/mpp.13017. Epub 2020 Nov 27. Mol Plant Pathol. 2021. PMID: 33245804 Free PMC article.
-
Delicate balance between functionally required flexibility and aggregation risk in a β-rich protein.Biochemistry. 2013 Dec 10;52(49):8843-54. doi: 10.1021/bi4013462. Epub 2013 Nov 25. Biochemistry. 2013. PMID: 24236614 Free PMC article.
-
Dicationic Imidazolium-Based Ionic Liquid Coatings on Zirconia Surfaces: Physico-Chemical and Biological Characterization.J Funct Biomater. 2017 Dec 13;8(4):50. doi: 10.3390/jfb8040050. J Funct Biomater. 2017. PMID: 29236088 Free PMC article.
-
Hydration of guanidinium depends on its local environment.Chem Sci. 2015 Jun 1;6(6):3420-3429. doi: 10.1039/c5sc00618j. Epub 2015 Apr 14. Chem Sci. 2015. PMID: 28706704 Free PMC article.
-
Aromatic clusters in protein-protein and protein-drug complexes.J Cheminform. 2020 May 8;12(1):30. doi: 10.1186/s13321-020-00437-4. J Cheminform. 2020. PMID: 33431014 Free PMC article.
References
-
- Waters ML. Aromatic interactions in model systems. Curr Opin Chem Biol. 2002;6:736–41. - PubMed
-
- Chakrabarti P, Bhattacharyya R. Geometry of nonbonded interactions involving planar groups in proteins. Prog Biophys Mol Biol. 2007;95:83–137. - PubMed
-
- Malkov SN, Zivkovic MV, Beljanski MV, Hall MB, Zaric SD. A reexamination of the propensities of amino acids towards a particular secondary structure: classification of amino acids based on their chemical structure. J Mol Model. 2008;14:769–75. - PubMed
-
- Thomas A, Meurisse R, Charloteaux B, Brasseur R. Aromatic side-chain interactions in proteins. I Main structural features. Proteins. 2002;48:628–34. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

