De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements
- PMID: 23811145
- PMCID: PMC3787258
- DOI: 10.1101/gr.157271.113
De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements
Abstract
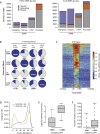
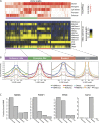
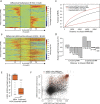
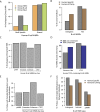
Deep sequencing of mammalian DNA methylomes has uncovered a previously unpredicted number of discrete hypomethylated regions in intergenic space (iHMRs). Here, we combined whole-genome bisulfite sequencing data with extensive gene expression and chromatin-state data to define functional classes of iHMRs, and to reconstruct the dynamics of their establishment in a developmental setting. Comparing HMR profiles in embryonic stem and primary blood cells, we show that iHMRs mark an exclusive subset of active DNase hypersensitive sites (DHS), and that both developmentally constitutive and cell-type-specific iHMRs display chromatin states typical of distinct regulatory elements. We also observe that iHMR changes are more predictive of nearby gene activity than the promoter HMR itself, and that expression of noncoding RNAs within the iHMR accompanies full activation and complete demethylation of mature B cell enhancers. Conserved sequence features corresponding to iHMR transcript start sites, including a discernible TATA motif, suggest a conserved, functional role for transcription in these regions. Similarly, we explored both primate-specific and human population variation at iHMRs, finding that while enhancer iHMRs are more variable in sequence and methylation status than any other functional class, conservation of the TATA box is highly predictive of iHMR maintenance, reflecting the impact of sequence plasticity and transcriptional signals on iHMR establishment. Overall, our analysis allowed us to construct a three-step timeline in which (1) intergenic DHS are pre-established in the stem cell, (2) partial demethylation of blood-specific intergenic DHSs occurs in blood progenitors, and (3) complete iHMR formation and transcription coincide with enhancer activation in lymphoid-specified cells.
Figures


References
-
- Arnosti DN, Kulkarni MM 2005. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
