Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy
- PMID: 23811281
- PMCID: PMC7114207
- DOI: 10.1016/j.antiviral.2013.06.009
Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy
Abstract
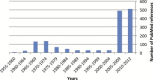
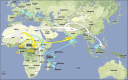
Chikungunya virus (CHIKV) is the aetiological agent of the mosquito-borne disease chikungunya fever, a debilitating arthritic disease that, during the past 7years, has caused immeasurable morbidity and some mortality in humans, including newborn babies, following its emergence and dispersal out of Africa to the Indian Ocean islands and Asia. Since the first reports of its existence in Africa in the 1950s, more than 1500 scientific publications on the different aspects of the disease and its causative agent have been produced. Analysis of these publications shows that, following a number of studies in the 1960s and 1970s, and in the absence of autochthonous cases in developed countries, the interest of the scientific community remained low. However, in 2005 chikungunya fever unexpectedly re-emerged in the form of devastating epidemics in and around the Indian Ocean. These outbreaks were associated with mutations in the viral genome that facilitated the replication of the virus in Aedes albopictus mosquitoes. Since then, nearly 1000 publications on chikungunya fever have been referenced in the PubMed database. This article provides a comprehensive review of chikungunya fever and CHIKV, including clinical data, epidemiological reports, therapeutic aspects and data relating to animal models for in vivo laboratory studies. It includes Supplementary Tables of all WHO outbreak bulletins, ProMED Mail alerts, viral sequences available on GenBank, and PubMed reports of clinical cases and seroprevalence studies.
Keywords: Alphavirus; Antiviral therapy; Arbovirus; Chikungunya fever; Chikungunya virus.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures







References
-
- Aikat B.K., Konar N.R., Banerjee G. Haemorrhagic Fever in Calcutta Area. Indian J. Med. Res. 1964;52:660–675. - PubMed
-
- Ali Ou Alla S., Combe B. Arthritis after infection with Chikungunya virus. Best Pract. Res. Clin. Rheumatol. 2011;25:337–346. - PubMed
-
- Allard P.M., Leyssen P., Martin M.T., Bourjot M., Dumontet V., Eydoux C., Guillemot J.C., Canard B., Poullain C., Gueritte F., Litaudon M. Antiviral chlorinated daphnane diterpenoid orthoesters from the bark and wood of Trigonostemon cherrieri. Phytochemistry. 2012;84:160–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

