pA506, a conjugative plasmid of the plant epiphyte Pseudomonas fluorescens A506
- PMID: 23811504
- PMCID: PMC3753976
- DOI: 10.1128/AEM.01354-13
pA506, a conjugative plasmid of the plant epiphyte Pseudomonas fluorescens A506
Abstract
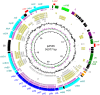
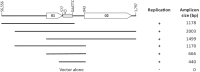
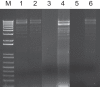
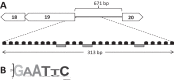
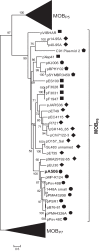
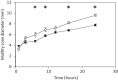
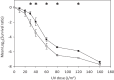
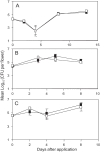
Conjugative plasmids are known to facilitate the acquisition and dispersal of genes contributing to the fitness of Pseudomonas spp. Here, we report the characterization of pA506, the 57-kb conjugative plasmid of Pseudomonas fluorescens A506, a plant epiphyte used in the United States for the biological control of fire blight disease of pear and apple. Twenty-nine of the 67 open reading frames (ORFs) of pA506 have putative functions in conjugation, including a type IV secretion system related to that of MOBP6 family plasmids and a gene cluster for type IV pili. We demonstrate that pA506 is self-transmissible via conjugation between A506 and strains of Pseudomonas spp. or the Enterobacteriaceae. The origin of vegetative replication (oriV) of pA506 is typical of those in pPT23A family plasmids, which are present in many pathovars of Pseudomonas syringae, but pA506 lacks repA, a defining locus for pPT23A plasmids, and has a novel partitioning region. We selected a plasmid-cured derivative of A506 and compared it to the wild type to identify plasmid-encoded phenotypes. pA506 conferred UV resistance, presumably due to the plasmid-borne rulAB genes, but did not influence epiphytic fitness of A506 on pear or apple blossoms in the field. pA506 does not appear to confer resistance to antibiotics or other toxic elements. Based on the conjugative nature of pA506 and the large number of its genes that are shared with plasmids from diverse groups of environmental bacteria, the plasmid is likely to serve as a vehicle for genetic exchange between A506 and its coinhabitants on plant surfaces.
Figures
Similar articles
-
Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family.BMC Genomics. 2017 May 10;18(1):365. doi: 10.1186/s12864-017-3763-x. BMC Genomics. 2017. PMID: 28486968 Free PMC article.
-
Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2.Mol Genet Genomics. 2004 Jan;270(6):462-76. doi: 10.1007/s00438-003-0945-9. Epub 2003 Nov 21. Mol Genet Genomics. 2004. PMID: 14634868
-
Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula.Phytopathology. 2010 Dec;100(12):1330-9. doi: 10.1094/PHYTO-03-10-0097. Phytopathology. 2010. PMID: 20839963
-
Stress tolerance and environmental fitness of Pseudomonas fluorescens A506, which has a mutation in RpoS.Phytopathology. 2009 Jun;99(6):679-88. doi: 10.1094/PHYTO-99-6-0679. Phytopathology. 2009. PMID: 19453226
-
Complete nucleotide sequence of the self-transmissible TOL plasmid pD2RT provides new insight into arrangement of toluene catabolic plasmids.Plasmid. 2013 Nov;70(3):393-405. doi: 10.1016/j.plasmid.2013.09.003. Epub 2013 Oct 2. Plasmid. 2013. PMID: 24095800
Cited by
-
Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family.BMC Genomics. 2017 May 10;18(1):365. doi: 10.1186/s12864-017-3763-x. BMC Genomics. 2017. PMID: 28486968 Free PMC article.
-
Complete sequence and detailed analysis of the first indigenous plasmid from Xanthomonas oryzae pv. oryzicola.BMC Microbiol. 2015 Oct 24;15:233. doi: 10.1186/s12866-015-0562-x. BMC Microbiol. 2015. PMID: 26498126 Free PMC article.
-
Certain Listeria monocytogenes plasmids contribute to increased UVC ultraviolet light stress.FEMS Microbiol Lett. 2021 Sep 22;368(17):fnab123. doi: 10.1093/femsle/fnab123. FEMS Microbiol Lett. 2021. PMID: 34498664 Free PMC article.
-
Plasmid Replicons from Pseudomonas Are Natural Chimeras of Functional, Exchangeable Modules.Front Microbiol. 2017 Feb 13;8:190. doi: 10.3389/fmicb.2017.00190. eCollection 2017. Front Microbiol. 2017. PMID: 28243228 Free PMC article.
-
Comparative genomics of Pseudomonas syringae pv. syringae strains B301D and HS191 and insights into intrapathovar traits associated with plant pathogenesis.Microbiologyopen. 2015 Aug;4(4):553-73. doi: 10.1002/mbo3.261. Epub 2015 May 4. Microbiologyopen. 2015. PMID: 25940918 Free PMC article.
References
-
- Fullbrook PD, Elson SW, Slocombe B. 1970. R-factor mediated beta-lactamase in Pseudomonas aeruginosa. Nature 226:1054–1056 - PubMed
-
- Gootz TD. 2010. The global problem of antibiotic resistance. Crit. Rev. Immunol. 30:79–93 - PubMed
-
- Sundin GW. 2007. Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 45:129. - PubMed
-
- Dennis JJ. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291–298 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

