Early phenylpropanoid biosynthetic steps in Cannabis sativa: link between genes and metabolites
- PMID: 23812081
- PMCID: PMC3742207
- DOI: 10.3390/ijms140713626
Early phenylpropanoid biosynthetic steps in Cannabis sativa: link between genes and metabolites
Abstract
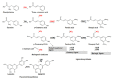
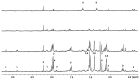
Phenylalanine ammonia-lyase (PAL), Cinnamic acid 4-hydroxylase (C4H) and 4-Coumarate: CoA ligase (4CL) catalyze the first three steps of the general phenylpropanoid pathway whereas chalcone synthase (CHS) catalyzes the first specific step towards flavonoids production. This class of specialized metabolites has a wide range of biological functions in plant development and defence and a broad spectrum of therapeutic activities for human health. In this study, we report the isolation of hemp PAL and 4CL cDNA and genomic clones. Through in silico analysis of their deduced amino acid sequences, more than an 80% identity with homologues genes of other plants was shown and phylogenetic relationships were highlighted. Quantitative expression analysis of the four above mentioned genes, PAL and 4CL enzymatic activities, lignin content and NMR metabolite fingerprinting in different Cannabis sativa tissues were evaluated. Furthermore, the use of different substrates to assay PAL and 4CL enzymatic activities indicated that different isoforms were active in different tissues. The diversity in secondary metabolites content observed in leaves (mainly flavonoids) and roots (mainly lignin) was discussed in relation to gene expression and enzymatic activities data.
Figures





References
-
- Flores-Sanchez I.J., Verpoorte R. Secondary metabolism in cannabis. Phytochem. Rev. 2008;7:615–639.
-
- Li X., Wang S., Du G., Wu Z., Meng Y. Variation in physical and mechanical properties of hemp stalk fibers along height of stem. Ind. Crops Prod. 2013;42:344–348.
-
- Ware M.A., Tawfik V.L. Safety issues concerning the medical use of cannabis and cannabinoids. Pain Res. Manag. 2005;10:31A–37A. - PubMed
-
- Jiang H.E., Li X., Zhao Y.X., Ferguson D.K., Hueber F., Bera S., Wang Y.F., Zhao L.C., Liu C.J., Li C.S. A new insight into Cannabis sativa (Cannabaceae) utilization from 2500-year-old Yanghai Tombs, Xinjiang, China. J. Ethnopharmacol. 2006;108:4014–4422. - PubMed
-
- Kostic M., Pejic B., Skundric P. Quality of chemically modified hemp fibres. Biorsource Technol. 2008;99:94–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

