Elongation factor G bound to the ribosome in an intermediate state of translocation
- PMID: 23812720
- PMCID: PMC3836249
- DOI: 10.1126/science.1235490
Elongation factor G bound to the ribosome in an intermediate state of translocation
Abstract
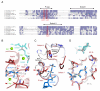
A key step of translation by the ribosome is translocation, which involves the movement of messenger RNA (mRNA) and transfer RNA (tRNA) with respect to the ribosome. This allows a new round of protein chain elongation by placing the next mRNA codon in the A site of the 30S subunit. Translocation proceeds through an intermediate state in which the acceptor ends of the tRNAs have moved with respect to the 50S subunit but not the 30S subunit, to form hybrid states. The guanosine triphosphatase (GTPase) elongation factor G (EF-G) catalyzes the subsequent movement of mRNA and tRNA with respect to the 30S subunit. Here, we present a crystal structure at 3 angstrom resolution of the Thermus thermophilus ribosome with a tRNA in the hybrid P/E state bound to EF-G with a GTP analog. The structure provides insights into structural changes that facilitate translocation and suggests a common GTPase mechanism for EF-G and elongation factor Tu.
Figures





Comment in
-
Biochemistry. Translocation in action.Science. 2013 Jun 28;340(6140):1534-5. doi: 10.1126/science.1240090. Science. 2013. PMID: 23812707 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

