Supervised de novo reconstruction of metabolic pathways from metabolome-scale compound sets
- PMID: 23812977
- PMCID: PMC3694648
- DOI: 10.1093/bioinformatics/btt244
Supervised de novo reconstruction of metabolic pathways from metabolome-scale compound sets
Abstract
Motivation: The metabolic pathway is an important biochemical reaction network involving enzymatic reactions among chemical compounds. However, it is assumed that a large number of metabolic pathways remain unknown, and many reactions are still missing even in known pathways. Therefore, the most important challenge in metabolomics is the automated de novo reconstruction of metabolic pathways, which includes the elucidation of previously unknown reactions to bridge the metabolic gaps.
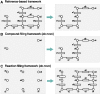
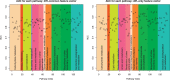
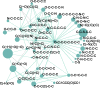
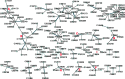
Results: In this article, we develop a novel method to reconstruct metabolic pathways from a large compound set in the reaction-filling framework. We define feature vectors representing the chemical transformation patterns of compound-compound pairs in enzymatic reactions using chemical fingerprints. We apply a sparsity-induced classifier to learn what we refer to as 'enzymatic-reaction likeness', i.e. whether compound pairs are possibly converted to each other by enzymatic reactions. The originality of our method lies in the search for potential reactions among many compounds at a time, in the extraction of reaction-related chemical transformation patterns and in the large-scale applicability owing to the computational efficiency. In the results, we demonstrate the usefulness of our proposed method on the de novo reconstruction of 134 metabolic pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG). Our comprehensively predicted reaction networks of 15 698 compounds enable us to suggest many potential pathways and to increase research productivity in metabolomics.
Availability: Softwares are available on request. Supplementary material are available at http://web.kuicr.kyoto-u.ac.jp/supp/kot/ismb2013/.
Figures

Similar articles
-
Simultaneous prediction of enzyme orthologs from chemical transformation patterns for de novo metabolic pathway reconstruction.Bioinformatics. 2016 Jun 15;32(12):i278-i287. doi: 10.1093/bioinformatics/btw260. Bioinformatics. 2016. PMID: 27307627 Free PMC article.
-
Metabolome-scale prediction of intermediate compounds in multistep metabolic pathways with a recursive supervised approach.Bioinformatics. 2014 Jun 15;30(12):i165-74. doi: 10.1093/bioinformatics/btu265. Bioinformatics. 2014. PMID: 24931980 Free PMC article.
-
Metabolome-scale de novo pathway reconstruction using regioisomer-sensitive graph alignments.Bioinformatics. 2015 Jun 15;31(12):i161-70. doi: 10.1093/bioinformatics/btv224. Bioinformatics. 2015. PMID: 26072478 Free PMC article.
-
Unveiling cellular biochemical reactions via metabolomics-driven approaches.Curr Opin Microbiol. 2010 Jun;13(3):358-62. doi: 10.1016/j.mib.2010.04.006. Epub 2010 Apr 27. Curr Opin Microbiol. 2010. PMID: 20430690 Review.
-
Analyzing methods for path mining with applications in metabolomics.Gene. 2014 Jan 25;534(2):125-38. doi: 10.1016/j.gene.2013.10.056. Epub 2013 Nov 12. Gene. 2014. PMID: 24230973 Review.
Cited by
-
Implementation and comparison of kernel-based learning methods to predict metabolic networks.Netw Model Anal Health Inform Bioinform. 2016;5(1):26. doi: 10.1007/s13721-016-0134-5. Epub 2016 Jul 15. Netw Model Anal Health Inform Bioinform. 2016. PMID: 27471658 Free PMC article.
-
Simultaneous prediction of enzyme orthologs from chemical transformation patterns for de novo metabolic pathway reconstruction.Bioinformatics. 2016 Jun 15;32(12):i278-i287. doi: 10.1093/bioinformatics/btw260. Bioinformatics. 2016. PMID: 27307627 Free PMC article.
-
Metabolome-scale prediction of intermediate compounds in multistep metabolic pathways with a recursive supervised approach.Bioinformatics. 2014 Jun 15;30(12):i165-74. doi: 10.1093/bioinformatics/btu265. Bioinformatics. 2014. PMID: 24931980 Free PMC article.
-
Metabolome-scale de novo pathway reconstruction using regioisomer-sensitive graph alignments.Bioinformatics. 2015 Jun 15;31(12):i161-70. doi: 10.1093/bioinformatics/btv224. Bioinformatics. 2015. PMID: 26072478 Free PMC article.
-
Dual graph convolutional neural network for predicting chemical networks.BMC Bioinformatics. 2020 Apr 23;21(Suppl 3):94. doi: 10.1186/s12859-020-3378-0. BMC Bioinformatics. 2020. PMID: 32321421 Free PMC article.
References
-
- Ben-Hur A, Noble W. Kernel methods for predicting protein–protein interactions. Bioinformatics. 2005;21(Suppl. 1):i38–i46. - PubMed
-
- Bono H, et al. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998;8:203–220. - PubMed
-
- Cascante M, et al. Metabolic control analysis in drug discovery and disease. Nat. Biotechnol. 2002;20:243–249. - PubMed
-
- Darvas F. Predicting metabolic pathways by logic programming. J. Mol. Graph. 1988;6:80–86.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

