Learning subgroup-specific regulatory interactions and regulator independence with PARADIGM
- PMID: 23813010
- PMCID: PMC3694636
- DOI: 10.1093/bioinformatics/btt229
Learning subgroup-specific regulatory interactions and regulator independence with PARADIGM
Abstract
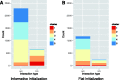
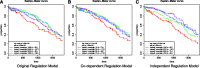
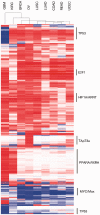
High-dimensional '-omics' profiling provides a detailed molecular view of individual cancers; however, understanding the mechanisms by which tumors evade cellular defenses requires deep knowledge of the underlying cellular pathways within each cancer sample. We extended the PARADIGM algorithm (Vaske et al., 2010, Bioinformatics, 26, i237-i245), a pathway analysis method for combining multiple '-omics' data types, to learn the strength and direction of 9139 gene and protein interactions curated from the literature. Using genomic and mRNA expression data from 1936 samples in The Cancer Genome Atlas (TCGA) cohort, we learned interactions that provided support for and relative strength of 7138 (78%) of the curated links. Gene set enrichment found that genes involved in the strongest interactions were significantly enriched for transcriptional regulation, apoptosis, cell cycle regulation and response to tumor cells. Within the TCGA breast cancer cohort, we assessed different interaction strengths between breast cancer subtypes, and found interactions associated with the MYC pathway and the ER alpha network to be among the most differential between basal and luminal A subtypes. PARADIGM with the Naive Bayesian assumption produced gene activity predictions that, when clustered, found groups of patients with better separation in survival than both the original version of PARADIGM and a version without the assumption. We found that this Naive Bayes assumption was valid for the vast majority of co-regulators, indicating that most co-regulators act independently on their shared target.
Availability: http://paradigm.five3genomics.com.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

