Integrating sequence, expression and interaction data to determine condition-specific miRNA regulation
- PMID: 23813013
- PMCID: PMC3694655
- DOI: 10.1093/bioinformatics/btt231
Integrating sequence, expression and interaction data to determine condition-specific miRNA regulation
Abstract
Motivation: MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression post-transcriptionally. MiRNAs were shown to play an important role in development and disease, and accurately determining the networks regulated by these miRNAs in a specific condition is of great interest. Early work on miRNA target prediction has focused on using static sequence information. More recently, researchers have combined sequence and expression data to identify such targets in various conditions.
Results: We developed the Protein Interaction-based MicroRNA Modules (PIMiM), a regression-based probabilistic method that integrates sequence, expression and interaction data to identify modules of mRNAs controlled by small sets of miRNAs. We formulate an optimization problem and develop a learning framework to determine the module regulation and membership. Applying PIMiM to cancer data, we show that by adding protein interaction data and modeling cooperative regulation of mRNAs by a small number of miRNAs, PIMiM can accurately identify both miRNA and their targets improving on previous methods. We next used PIMiM to jointly analyze a number of different types of cancers and identified both common and cancer-type-specific miRNA regulators.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures


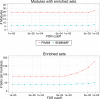

 modules learned for the three cancer types (each x-axis bar is subdivided into three with the color corresponding to the cancer type). The y-axis shows miRNAs ordered by hierarchical clustering of their module membership vector. In several cases, the same miRNAs are predicted for all or two of the three cancer types
modules learned for the three cancer types (each x-axis bar is subdivided into three with the color corresponding to the cancer type). The y-axis shows miRNAs ordered by hierarchical clustering of their module membership vector. In several cases, the same miRNAs are predicted for all or two of the three cancer types
Similar articles
-
Tiresias: Context-sensitive Approach to Decipher the Presence and Strength of MicroRNA Regulatory Interactions.Theranostics. 2018 Jan 1;8(1):277-291. doi: 10.7150/thno.22065. eCollection 2018. Theranostics. 2018. PMID: 29290807 Free PMC article.
-
Identifying functional miRNA-mRNA regulatory modules with correspondence latent dirichlet allocation.Bioinformatics. 2010 Dec 15;26(24):3105-11. doi: 10.1093/bioinformatics/btq576. Epub 2010 Oct 17. Bioinformatics. 2010. PMID: 20956247 Free PMC article.
-
LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer.Bioinformatics. 2018 Dec 15;34(24):4232-4240. doi: 10.1093/bioinformatics/bty525. Bioinformatics. 2018. PMID: 29955818
-
Disruption of microRNA nuclear transport in human cancer.Semin Cancer Biol. 2014 Aug;27:46-51. doi: 10.1016/j.semcancer.2014.02.012. Epub 2014 Mar 7. Semin Cancer Biol. 2014. PMID: 24607282 Review.
-
The miRNA-kallikrein interaction: a mosaic of epigenetic regulation in cancer.Biol Chem. 2018 Sep 25;399(9):973-982. doi: 10.1515/hsz-2018-0112. Biol Chem. 2018. PMID: 29604203 Review.
Cited by
-
Integrative analysis of multi-omics data for identifying multi-markers for diagnosing pancreatic cancer.BMC Genomics. 2015;16 Suppl 9(Suppl 9):S4. doi: 10.1186/1471-2164-16-S9-S4. Epub 2015 Aug 17. BMC Genomics. 2015. PMID: 26328610 Free PMC article.
-
Inferring condition-specific miRNA activity from matched miRNA and mRNA expression data.Bioinformatics. 2014 Nov 1;30(21):3070-7. doi: 10.1093/bioinformatics/btu489. Epub 2014 Jul 23. Bioinformatics. 2014. PMID: 25061069 Free PMC article.
-
Gene-microRNA network module analysis for ovarian cancer.BMC Syst Biol. 2016 Dec 23;10(Suppl 4):117. doi: 10.1186/s12918-016-0357-1. BMC Syst Biol. 2016. PMID: 28155675 Free PMC article.
-
Connecting rules from paired miRNA and mRNA expression data sets of HCV patients to detect both inverse and positive regulatory relationships.BMC Genomics. 2015;16 Suppl 2(Suppl 2):S11. doi: 10.1186/1471-2164-16-S2-S11. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25707620 Free PMC article.
-
Identification of miRNA-mRNA regulatory modules by exploring collective group relationships.BMC Genomics. 2016 Jan 11;17 Suppl 1(Suppl 1):7. doi: 10.1186/s12864-015-2300-z. BMC Genomics. 2016. PMID: 26817421 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

