Integrating peptides' sequence and energy of contact residues information improves prediction of peptide and HLA-I binding with unknown alleles
- PMID: 23815611
- PMCID: PMC3654895
- DOI: 10.1186/1471-2105-14-S8-S1
Integrating peptides' sequence and energy of contact residues information improves prediction of peptide and HLA-I binding with unknown alleles
Abstract
Background: The HLA (human leukocyte antigen) class I is a kind of molecule encoded by a large family of genes and is characteristic of high polymorphism. Now the number of the registered HLA-I molecules has exceeded 3000. Slight differences in the amino acid sequences of HLAs would make them bind to different sets of peptides. In the past decades, although many methods have been proposed to predict the binding between peptides and HLA-I molecules and achieved good performance, most experimental data used by them is limited to the HLAs with a small number of alleles. Thus they are inclined to obtain high prediction accuracy only for data with similar alleles. Because the peptides and HLAs together determine the binding, it's necessary to consider their contribution meanwhile.
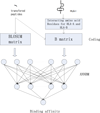
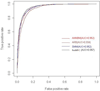
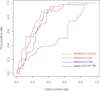
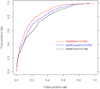
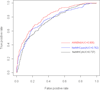
Results: By taking into account the features of the peptides sequence and the energy of contact residues, in this paper a method based on the artificial neural network is proposed to predict the binding of peptides and HLA-I even when the HLAs' potential alleles are unknown. Two experiments in the allele-specific and super-type cases are performed respectively to validate our method. In the first case, we collect 14 HLA-A and 14 HLA-B molecules on Bjoern Peters dataset, and compare our method with the ARB, SMM, NetMHC and other 16 online methods. Our method gets the best average AUC (Area under the ROC) value as 0.909. In the second one, we use leave one out cross validation on MHC-peptide binding data that has different alleles but shares the common super-type. Compared to gold standard methods like NetMHC and NetMHCpan, our method again achieves the best average AUC value as 0.847.
Conclusions: Our method achieves satisfactory results. Whenever it's tested on the HLA-I with single definite gene or with super-type gene locus, it gets better classification accuracy. Especially, when the training set is small, our method still works better than the other methods in the comparison. Therefore, we could make a conclusion that by combining the peptides' information, HLAs amino acid residues' interaction information and contact energy, our method really could improve prediction of the peptide HLA-I binding even when there aren't the prior experimental dataset for HLAs with various alleles.
Figures

Similar articles
-
NetMHCpan, a method for MHC class I binding prediction beyond humans.Immunogenetics. 2009 Jan;61(1):1-13. doi: 10.1007/s00251-008-0341-z. Epub 2008 Nov 12. Immunogenetics. 2009. PMID: 19002680 Free PMC article.
-
DeepSeqPan, a novel deep convolutional neural network model for pan-specific class I HLA-peptide binding affinity prediction.Sci Rep. 2019 Jan 28;9(1):794. doi: 10.1038/s41598-018-37214-1. Sci Rep. 2019. PMID: 30692623 Free PMC article.
-
Understanding and predicting binding between human leukocyte antigens (HLAs) and peptides by network analysis.BMC Bioinformatics. 2015;16 Suppl 13(Suppl 13):S9. doi: 10.1186/1471-2105-16-S13-S9. Epub 2015 Sep 25. BMC Bioinformatics. 2015. PMID: 26424483 Free PMC article.
-
A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction.Brief Bioinform. 2020 Jul 15;21(4):1119-1135. doi: 10.1093/bib/bbz051. Brief Bioinform. 2020. PMID: 31204427 Free PMC article. Review.
-
Class I MHC-peptide interaction: structural and functional aspects.Behring Inst Mitt. 1994 Jul;(94):48-60. Behring Inst Mitt. 1994. PMID: 7998914 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

