A multi-site feasibility study for personalized medicine in canines with osteosarcoma
- PMID: 23815880
- PMCID: PMC3702405
- DOI: 10.1186/1479-5876-11-158
A multi-site feasibility study for personalized medicine in canines with osteosarcoma
Abstract
Background: A successful therapeutic strategy, specifically tailored to the molecular constitution of an individual and their disease, is an ambitious objective of modern medicine. In this report, we highlight a feasibility study in canine osteosarcoma focused on refining the infrastructure and processes required for prospective clinical trials using a series of gene expression-based Personalized Medicine (PMed) algorithms to predict suitable therapies within 5 days of sample receipt.
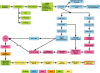
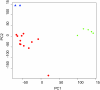
Methods: Tumor tissue samples were collected immediately following limb amputation and shipped overnight from veterinary practices. Upon receipt (day 1), RNA was extracted from snap-frozen tissue, with an adjacent H&E section for pathological diagnosis. Samples passing RNA and pathology QC were shipped to a CLIA-certified laboratory for genomic profiling. After mapping of canine probe sets to human genes and normalization against a (normal) reference set, gene level Z-scores were submitted to the PMed algorithms. The resulting PMed report was immediately forwarded to the veterinarians. Upon receipt and review of the PMed report, feedback from the practicing veterinarians was captured.
Results: 20 subjects were enrolled over a 5 month period. Tissue from 13 subjects passed both histological and RNA QC and were submitted for genomic analysis and subsequent PMed analysis and report generation. 11 of the 13 samples for which PMed reports were produced were communicated to the veterinarian within the target 5 business days. Of the 7 samples that failed QC, 4 were due to poor RNA quality, whereas 2 were failed following pathological review. Comments from the practicing veterinarians were generally positive and constructive, highlighting a number of areas for improvement, including enhanced education regarding PMed report interpretation, drug availability, affordable pricing and suitable canine dosing.
Conclusions: This feasibility trial demonstrated that with the appropriate infrastructure and processes it is possible to perform an in-depth molecular analysis of a patient's tumor in support of real time therapeutic decision making within 5 days of sample receipt. A number of areas for improvement have been identified that should reduce the level of sample attrition and support clinical decision making.
Figures

References
-
- Gasparini G, Longo R. The paradigm of personalized therapy in oncology. Expert Opin Ther Targets. 2012;16(Suppl 1):S7–S16. - PubMed
-
- Blay JY, Le Cesne A, Cassier PA, Ray-Coquard IL. Gastrointestinal Stromal Tumors (GIST): a rare entity, a tumor model for personalized therapy, and yet ten different molecular subtypes. Discov Med. 2012;13:357–367. - PubMed
-
- Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127. ps110. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

