Emerging regulation and functions of autophagy
- PMID: 23817233
- PMCID: PMC7097732
- DOI: 10.1038/ncb2788
Emerging regulation and functions of autophagy
Erratum in
- Nat Cell Biol. 2013 Aug;15(8):1017
Abstract
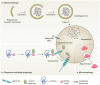
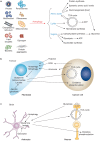
Autophagy maintains cell, tissue and organism homeostasis through degradation. Complex post-translational modulation of the Atg (autophagy-related) proteins adds additional entry points for crosstalk with other cellular processes and helps define cell-type-specific regulations of autophagy. Beyond the simplistic view of a process exclusively dedicated to the turnover of cellular components, recent data have uncovered unexpected functions for autophagy and the autophagy-related genes, such as regulation of metabolism, membrane transport and modulation of host defenses--indicating the novel frontiers lying ahead.
Figures
References
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. - PubMed
-
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012;22:R29–34. - PubMed
-
- Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

