Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii
- PMID: 23818097
- PMCID: PMC3776330
- DOI: 10.1002/pro.2302
Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii
Abstract
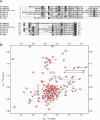
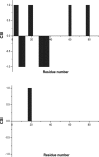
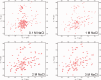
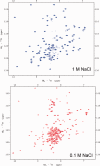
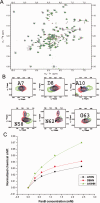
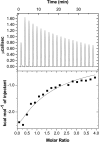
Eukaryotic ubiquitin and ubiquitin-like systems play crucial roles in various cellular biological processes. In this work, we determined the solution structure of SAMP1 from Haloferax volcanii by NMR spectroscopy. Under low ionic conditions, SAMP1 presented two distinct conformations, one folded β-grasp and the other disordered. Interestingly, SAMP1 underwent a conformational conversion from disorder to order with ion concentration increasing, indicating that the ordered conformation is the functional form of SAMP1 under the physiological condition of H. volcanii. Furthermore, SAMP1 could interact with proteasome-activating nucleotidase B, supposing a potential role of SAMP1 in the protein degradation pathway mediated by proteasome.
Keywords: Haloferax volcanii; NMR; SAMP1; protein folding; ubiquitin-like protein.
© 2013 The Protein Society.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

