Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing
- PMID: 23818871
- PMCID: PMC3688494
- DOI: 10.1371/journal.pgen.1003584
Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing
Abstract
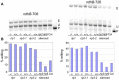
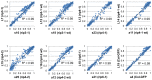
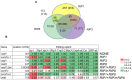
In flowering plants, mitochondrial and chloroplast mRNAs are edited by C-to-U base modification. In plant organelles, RNA editing appears to be generally a correcting mechanism that restores the proper function of the encoded product. Members of the Arabidopsis RNA editing-Interacting Protein (RIP) family have been recently shown to be essential components of the plant editing machinery. We report the use of a strand- and transcript-specific RNA-seq method (STS-PCRseq) to explore the effect of mutation or silencing of every RIP gene on plant organelle editing. We confirm RIP1 to be a major editing factor that controls the editing extent of 75% of the mitochondrial sites and 20% of the plastid C targets of editing. The quantitative nature of RNA sequencing allows the precise determination of overlapping effects of RIP factors on RNA editing. Over 85% of the sites under the influence of RIP3 and RIP8, two moderately important mitochondrial factors, are also controlled by RIP1. Previously uncharacterized RIP family members were found to have only a slight effect on RNA editing. The preferential location of editing sites controlled by RIP7 on some transcripts suggests an RNA metabolism function for this factor other than editing. In addition to a complete characterization of the RIP factors for their effect on RNA editing, our study highlights the potential of RNA-seq for studying plant organelle editing. Unlike previous attempts to use RNA-seq to analyze RNA editing extent, our methodology focuses on sequencing of organelle cDNAs corresponding to known transcripts. As a result, the depth of coverage of each editing site reaches unprecedented values, assuring a reliable measurement of editing extent and the detection of numerous new sites. This strategy can be applied to the study of RNA editing in any organism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Gott JM, Emeson RB (2000) Functions and mechanisms of RNA editing. Annu Rev Genet 34: 499–531. - PubMed
-
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-Garcia A, Andres C, de la Luz Gutierrez-Nava M, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602. - PubMed
-
- Staudinger M, Kempken F (2003) Electroporation of isolated higher-plant mitochondria: Transcripts of an introduced cox2 gene, but not an atp6 gene, are edited in organello . Mol Genet Genomics 269: 553–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

