Blood Clotting Factor VIII: From Evolution to Therapy
- PMID: 23819034
- PMCID: PMC3695351
Blood Clotting Factor VIII: From Evolution to Therapy
Abstract
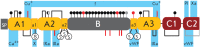
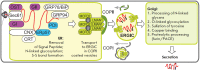
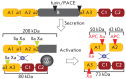
Recombinant blood clotting factor VIII is one of the most complex proteins for industrial manufacturing due to the low efficiency of its gene transcription, massive intracellular loss of its proprotein during post-translational processing, and the instability of the secreted protein. Improvement in hemophilia A therapy requires a steady increase in the production of factor VIII drugs despite tightening standards of product quality and viral safety. More efficient systems for heterologous expression of factor VIII can be created on the basis of the discovered properties of its gene transcription, post-translational processing, and behavior in the bloodstream. The present review describes the deletion variants of factor VIII protein with increased secretion efficiency and the prospects for the pharmaceutical development of longer acting variants and derivatives of factor VIII.
Keywords: blood clotting factor VIII; hemophilia A; heterologous protein expression systems.
Figures

References
-
- Antonarakis S.E., Kazazian H.H., Tuddenham E.G.. Hum. Mutat. 1995;5(1):1–22. - PubMed
-
- A Mutation Database. http://hadb.org.uk. Kemball-Cook G., Haemophilia A., University College London. 1994
-
- Blumel J., Schmidt I., Effenberger W., Seitz H., Willkommen H., Brackmann H.H., Lower J., Eis-Hubinger A.M.. Transfusion. 2002;42(11):1473–1481. - PubMed
-
- Yokozaki S., Fukuda Y., Nakano I., Katano Y., Toyoda H., Takamatsu J.. Blood. 1999;94(10):3617. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
