Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation
- PMID: 23819640
- PMCID: PMC3707822
- DOI: 10.1186/1756-8935-6-19
Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation
Abstract
Background: Smchd1 is an epigenetic modifier essential for X chromosome inactivation: female embryos lacking Smchd1 fail during midgestational development. Male mice are less affected by Smchd1-loss, with some (but not all) surviving to become fertile adults on the FVB/n genetic background. On other genetic backgrounds, all males lacking Smchd1 die perinatally. This suggests that, in addition to being critical for X inactivation, Smchd1 functions to control the expression of essential autosomal genes.
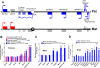
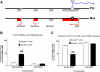
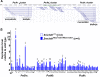
Results: Using genome-wide microarray expression profiling and RNA-seq, we have identified additional genes that fail X inactivation in female Smchd1 mutants and have identified autosomal genes in male mice where the normal expression pattern depends upon Smchd1. A subset of genes in the Snrpn imprinted gene cluster show an epigenetic signature and biallelic expression consistent with loss of imprinting in the absence of Smchd1. In addition, single nucleotide polymorphism analysis of expressed genes in the placenta shows that the Igf2r imprinted gene cluster is also disrupted, with Slc22a3 showing biallelic expression in the absence of Smchd1. In both cases, the disruption was not due to loss of the differential methylation that marks the imprint control region, but affected genes remote from this primary imprint controlling element. The clustered protocadherins (Pcdhα, Pcdhβ, and Pcdhγ) also show altered expression levels, suggesting that their unique pattern of random combinatorial monoallelic expression might also be disrupted.
Conclusions: Smchd1 has a role in the expression of several autosomal gene clusters that are subject to monoallelic expression, rather than being restricted to functioning uniquely in X inactivation. Our findings, combined with the recent report implicating heterozygous mutations of SMCHD1 as a causal factor in the digenically inherited muscular weakness syndrome facioscapulohumeral muscular dystrophy-2, highlight the potential importance of Smchd1 in the etiology of diverse human diseases.
Figures

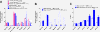

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

