Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging
- PMID: 23819996
- PMCID: PMC3748192
- DOI: 10.4161/auto.24811
Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging
Abstract
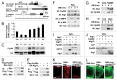
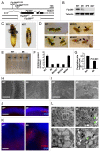
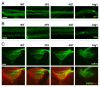
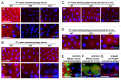
Autophagy-related 1 (Atg1)/Unc-51-like protein kinases (ULKs) are evolutionarily conserved proteins that play critical physiological roles in controlling autophagy, cell growth and neurodevelopment. RB1-inducible coiled-coil 1 (RB1CC1), also known as PTK2/FAK family-interacting protein of 200 kDa (FIP200) is a recently discovered binding partner of ULK1. Here we isolated the Drosophila RB1CC1/FIP200 homolog (Fip200/CG1347) and showed that it mediates Atg1-induced autophagy as a genetically downstream component in diverse physiological contexts. Fip200 loss-of-function mutants experienced severe mobility loss associated with neuronal autophagy defects and neurodegeneration. The Fip200 mutants were also devoid of both developmental and starvation-induced autophagy in salivary gland and fat body, while having no defects in axonal transport and projection in developing neurons. Interestingly, moderate downregulation of Fip200 accelerated both developmental growth and aging, accompanied by target of rapamycin (Tor) signaling upregulation. These results suggest that Fip200 is a critical downstream component of Atg1 and specifically mediates Atg1's autophagy-, aging- and growth-regulating functions.
Keywords: Drosophila; aging; autophagy; growth; neurodegeneration.
Figures


Similar articles
-
Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila.Autophagy. 2014 Mar;10(3):453-67. doi: 10.4161/auto.27442. Epub 2014 Jan 6. Autophagy. 2014. PMID: 24419107 Free PMC article.
-
The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway.Autophagy. 2018;14(5):796-811. doi: 10.1080/15548627.2017.1386820. Epub 2017 Dec 24. Autophagy. 2018. PMID: 29099309 Free PMC article.
-
ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase.EMBO Rep. 2007 Apr;8(4):360-5. doi: 10.1038/sj.embor.7400917. Epub 2007 Mar 9. EMBO Rep. 2007. PMID: 17347671 Free PMC article.
-
UNC51-like kinase 1, autophagic regulator and cancer therapeutic target.Cell Prolif. 2014 Dec;47(6):494-505. doi: 10.1111/cpr.12145. Epub 2014 Oct 20. Cell Prolif. 2014. PMID: 25327638 Free PMC article. Review.
-
Canonical and noncanonical functions of ULK/Atg1.Curr Opin Cell Biol. 2017 Apr;45:47-54. doi: 10.1016/j.ceb.2017.02.011. Epub 2017 Mar 11. Curr Opin Cell Biol. 2017. PMID: 28292700 Free PMC article. Review.
Cited by
-
Identification of an AMPK phosphorylation site in Drosophila TSC2 (gigas) that regulate cell growth.Int J Mol Sci. 2015 Mar 27;16(4):7015-26. doi: 10.3390/ijms16047015. Int J Mol Sci. 2015. PMID: 25826530 Free PMC article.
-
Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages.PLoS Pathog. 2014 Jun 5;10(6):e1004159. doi: 10.1371/journal.ppat.1004159. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24901456 Free PMC article.
-
The Drosophila ZNRF1/2 homologue, detour, interacts with HOPS complex and regulates autophagy.Commun Biol. 2024 Feb 15;7(1):183. doi: 10.1038/s42003-024-05834-1. Commun Biol. 2024. PMID: 38360932 Free PMC article.
-
Microarray Analysis of the Major Depressive Disorder mRNA Profile Data.Psychiatry Investig. 2015 Jul;12(3):388-96. doi: 10.4306/pi.2015.12.3.388. Epub 2015 Jul 6. Psychiatry Investig. 2015. PMID: 26207134 Free PMC article.
-
Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1.FEBS J. 2014 Sep;281(17):3816-27. doi: 10.1111/febs.12905. Epub 2014 Jul 28. FEBS J. 2014. PMID: 25040165 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30-DK034933/DK/NIDDK NIH HHS/United States
- P30-CA46592/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 AG013283/AG/NIA NIH HHS/United States
- P30-AG013283/AG/NIA NIH HHS/United States
- P30 AG024824/AG/NIA NIH HHS/United States
- R01 GM052890/GM/NIGMS NIH HHS/United States
- P30-AG024824/AG/NIA NIH HHS/United States
- P30 DK034933/DK/NIDDK NIH HHS/United States
- GM084947/GM/NIGMS NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R01-GM052890/GM/NIGMS NIH HHS/United States
- R01 GM084947/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
