Maximizing peptide identification events in proteomic workflows using data-dependent acquisition (DDA)
- PMID: 23820513
- PMCID: PMC3879624
- DOI: 10.1074/mcp.M112.026500
Maximizing peptide identification events in proteomic workflows using data-dependent acquisition (DDA)
Abstract
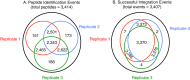
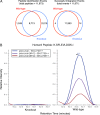
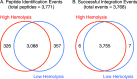
Current analytical strategies for collecting proteomic data using data-dependent acquisition (DDA) are limited by the low analytical reproducibility of the method. Proteomic discovery efforts that exploit the benefits of DDA, such as providing peptide sequence information, but that enable improved analytical reproducibility, represent an ideal scenario for maximizing measureable peptide identifications in "shotgun"-type proteomic studies. Therefore, we propose an analytical workflow combining DDA with retention time aligned extracted ion chromatogram (XIC) areas obtained from high mass accuracy MS1 data acquired in parallel. We applied this workflow to the analyses of sample matrixes prepared from mouse blood plasma and brain tissues and observed increases in peptide detection of up to 30.5% due to the comparison of peptide MS1 XIC areas following retention time alignment of co-identified peptides. Furthermore, we show that the approach is quantitative using peptide standards diluted into a complex matrix. These data revealed that peptide MS1 XIC areas provide linear response of over three orders of magnitude down to low femtomole (fmol) levels. These findings argue that augmenting "shotgun" proteomic workflows with retention time alignment of peptide identifications and comparative analyses of corresponding peptide MS1 XIC areas improve the analytical performance of global proteomic discovery methods using DDA.
Figures


References
-
- Wu C. C., MacCoss M. J. (2002) Shotgun proteomics: tools for the analysis of complex biological systems. Curr. Opin. Mol. Ther. 4, 242–250 - PubMed
-
- Neilson K. A., Ali N. A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S. C., Haynes P. A. (2011) Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 - PubMed
-
- Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 - PubMed
-
- Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 - PMC - PubMed
-
- Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

