The organization of the quorum sensing luxI/R family genes in Burkholderia
- PMID: 23820583
- PMCID: PMC3742214
- DOI: 10.3390/ijms140713727
The organization of the quorum sensing luxI/R family genes in Burkholderia
Abstract
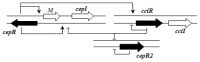
Members of the Burkholderia genus of Proteobacteria are capable of living freely in the environment and can also colonize human, animal and plant hosts. Certain members are considered to be clinically important from both medical and veterinary perspectives and furthermore may be important modulators of the rhizosphere. Quorum sensing via N-acyl homoserine lactone signals (AHL QS) is present in almost all Burkholderia species and is thought to play important roles in lifestyle changes such as colonization and niche invasion. Here we present a census of AHL QS genes retrieved from public databases and indicate that the local arrangement (topology) of QS genes, their location within chromosomes and their gene neighborhoods show characteristic patterns that differ between the known Burkholderia clades. In sequence phylogenies, AHL QS genes seem to cluster according to the local gene topology rather than according to the species, which suggests that the basic topology types were present prior to the appearance of current Burkholderia species. The data are available at http://net.icgeb.org/burkholderia/.
Figures







References
-
- Palleroni N.J., Kunisawa R., Contopoulou R., Doudoroff M. Nucleic Acid Homologies in the Genus Pseudomonas. Int. J. Syst. Bacteriol. 1973;23:333–339.
-
- Gillis M., Van T., Bardin R., Goor M., Hebbar P., Willems A., Segers P., Kersters K., Heulin T., Fernandez M.P. Polyphasic Taxonomy in the Genus Burkholderia Leading to an Emended Description of the Genus and Proposition of Burkholderia vietnamiensis sp. nov. for N2-Fixing Isolates from Rice in Vietnam. Int. J. Syst. Bacteriol. 1995;45:274–289.
-
- Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., Ezaki T., Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992;36:1251–1275. - PubMed
-
- Isles A., Maclusky I., Corey M., Gold R., Prober C., Fleming P., Levison H. Pseudomonas cepacia infection in cystic fibrosis: An emerging problem. J. Pediatr. 1984;104:206–210. - PubMed
-
- Vanlaere E., Lipuma J.J., Baldwin A., Henry D., De Brandt E., Mahenthiralingam E., Speert D., Dowson C., Vandamme P. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evolut. Microbiol. 2008;58:1580–1590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

