Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium
- PMID: 23821437
- PMCID: PMC7108517
- DOI: 10.1111/2049-632X.12053
Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium
Abstract
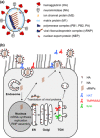
Influenza is an acute infection of the respiratory tract, which affects each year millions of people. Influenza virus infection is initiated by the surface glycoprotein hemagglutinin (HA) through receptor binding and fusion of viral and endosomal membranes. HA is synthesized as a precursor protein and requires cleavage by host cell proteases to gain its fusion capacity. Although cleavage of HA is crucial for virus infectivity, little was known about relevant proteases in the human airways for a long time. Recent progress in the identification and characterization of HA-activating host cell proteases has been considerable however and supports the idea of targeting HA cleavage as a novel approach for influenza treatment. Interestingly, certain bacteria have been demonstrated to support HA activation either by secreting proteases that cleave HA or due to activation of cellular proteases and thereby may contribute to virus spread and enhanced pathogenicity. In this review, we give an overview on activation of influenza viruses by proteases from host cells and bacteria with the main focus on recent progress on HA cleavage by proteases HAT and TMPRSS2 in the human airway epithelium. In addition, we outline investigations of HA-activating proteases as potential drug targets for influenza treatment.
Keywords: HAT; TMPRSS2; influenza treatment by protease inhibitors; influenza virus hemagglutinin; proteolytic cleavage; viral-bacterial pneumonia.
© 2013 Federation of European Microbiological Societies. Published by John Wiley & Sons Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Hemagglutinins of Avian Influenza Viruses Are Proteolytically Activated by TMPRSS2 in Human and Murine Airway Cells.J Virol. 2021 Sep 27;95(20):e0090621. doi: 10.1128/JVI.00906-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319155 Free PMC article.
-
TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes.J Virol. 2019 Oct 15;93(21):e00649-19. doi: 10.1128/JVI.00649-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31391268 Free PMC article.
-
Novel insights into proteolytic cleavage of influenza virus hemagglutinin.Rev Med Virol. 2010 Sep;20(5):298-310. doi: 10.1002/rmv.657. Rev Med Virol. 2010. PMID: 20629046 Free PMC article. Review.
-
Influenza virus activating host proteases: Identification, localization and inhibitors as potential therapeutics.Eur J Cell Biol. 2015 Jul-Sep;94(7-9):375-83. doi: 10.1016/j.ejcb.2015.05.013. Epub 2015 Jun 1. Eur J Cell Biol. 2015. PMID: 26095298
-
[The role of cleavage activation of the hemagglutinin by host and bacterial proteases in the induction of the pathogenesis of influenza viruses].Nihon Rinsho. 1997 Oct;55(10):2633-9. Nihon Rinsho. 1997. PMID: 9360383 Review. Japanese.
Cited by
-
Crystal structure of inhibitor-bound human MSPL that can activate high pathogenic avian influenza.Life Sci Alliance. 2021 Apr 5;4(6):e202000849. doi: 10.26508/lsa.202000849. Print 2021 Jun. Life Sci Alliance. 2021. PMID: 33820827 Free PMC article.
-
Recent Occurrence, Diversity, and Candidate Vaccine Virus Selection for Pandemic H5N1: Alert Is in the Air.Vaccines (Basel). 2024 Sep 12;12(9):1044. doi: 10.3390/vaccines12091044. Vaccines (Basel). 2024. PMID: 39340074 Free PMC article. Review.
-
Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens.Viruses. 2023 Feb 7;15(2):461. doi: 10.3390/v15020461. Viruses. 2023. PMID: 36851676 Free PMC article.
-
Port d'Entrée for Respiratory Infections - Does the Influenza A Virus Pave the Way for Bacteria?Front Microbiol. 2017 Dec 21;8:2602. doi: 10.3389/fmicb.2017.02602. eCollection 2017. Front Microbiol. 2017. PMID: 29312268 Free PMC article. Review.
-
Surface glycoprotein of Borna disease virus mediates virus spread from cell to cell.Cell Microbiol. 2016 Mar;18(3):340-54. doi: 10.1111/cmi.12515. Epub 2015 Oct 12. Cell Microbiol. 2016. PMID: 26332529 Free PMC article.
References
-
- Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB & Jakobovits A (2001) Catalytic cleavage of the androgen‐regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res 61: 1686–1692. - PubMed
-
- Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S & Soilleux EJ (2012) Influenza and SARS‐coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE 7: e35876. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

