Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition
- PMID: 23821663
- PMCID: PMC3783173
- DOI: 10.1093/nar/gkt590
Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition
Abstract
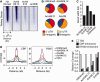
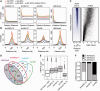
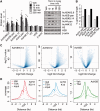
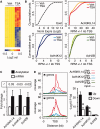
Histone deacetylase inhibitors (HDACis) have been shown to potentiate hippocampal-dependent memory and synaptic plasticity and to ameliorate cognitive deficits and degeneration in animal models for different neuropsychiatric conditions. However, the impact of these drugs on hippocampal histone acetylation and gene expression profiles at the genomic level, and the molecular mechanisms that underlie their specificity and beneficial effects in neural tissue, remains obscure. Here, we mapped four relevant histone marks (H3K4me3, AcH3K9,14, AcH4K12 and pan-AcH2B) in hippocampal chromatin and investigated at the whole-genome level the impact of HDAC inhibition on acetylation profiles and basal and activity-driven gene expression. HDAC inhibition caused a dramatic histone hyperacetylation that was largely restricted to active loci pre-marked with H3K4me3 and AcH3K9,14. In addition, the comparison of Chromatin immunoprecipitation sequencing and gene expression profiles indicated that Trichostatin A-induced histone hyperacetylation, like histone hypoacetylation induced by histone acetyltransferase deficiency, had a modest impact on hippocampal gene expression and did not affect the transient transcriptional response to novelty exposure. However, HDAC inhibition caused the rapid induction of a homeostatic gene program related to chromatin deacetylation. These results illuminate both the relationship between hippocampal gene expression and histone acetylation and the mechanism of action of these important neuropsychiatric drugs.
Figures


References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. - PubMed
-
- Lopez-Atalaya JP, Gervasini C, Mottadelli F, Spena S, Piccione M, Scarano G, Selicorni A, Barco A, Larizza L. Histone acetylation deficits in lymphoblastoid cell lines from patients with Rubinstein-Taybi syndrome. J. Med. Genet. 2012;49:66–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

