A germline point mutation in Runx1 uncouples its role in definitive hematopoiesis from differentiation
- PMID: 23823022
- PMCID: PMC3909992
- DOI: 10.1016/j.exphem.2013.06.006
A germline point mutation in Runx1 uncouples its role in definitive hematopoiesis from differentiation
Abstract
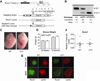
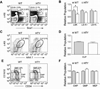
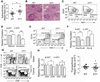
Definitive hematopoiesis requires the master hematopoietic transcription factor Runx1, which is a frequent target of leukemia-related chromosomal translocations. Several of the translocation-generated fusion proteins retain the DNA binding activity of Runx1, but lose subnuclear targeting and associated transactivation potential. Complete loss of these functions in vivo resembles Runx1 ablation, which causes embryonic lethality. We developed a knock-in mouse that expresses full-length Runx1 with a mutation in the subnuclear targeting cofactor interaction domain, Runx1(HTY350-352AAA). Mutant mice survive to adulthood, and hematopoietic stem cell emergence appears to be unaltered. However, defects are observed in multiple differentiated hematopoietic lineages at stages where Runx1 is known to play key roles. Thus, a germline mutation in Runx1 reveals uncoupling of its functions during developmental hematopoiesis from subsequent differentiation across multiple hematopoietic lineages in the adult. These findings indicate that subnuclear targeting and cofactor interactions with Runx1 are important in many compartments throughout hematopoietic differentiation.
Copyright © 2013 ISEH - Society for Hematology and Stem Cells. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures



Similar articles
-
Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation.Hum Mol Genet. 2010 Mar 15;19(6):1048-57. doi: 10.1093/hmg/ddp568. Epub 2009 Dec 24. Hum Mol Genet. 2010. PMID: 20035012 Free PMC article.
-
Ectopic Runx1 expression rescues Tal-1-deficiency in the generation of primitive and definitive hematopoiesis.PLoS One. 2013 Jul 29;8(7):e70116. doi: 10.1371/journal.pone.0070116. Print 2013. PLoS One. 2013. PMID: 23922928 Free PMC article.
-
The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis.Blood. 2009 Dec 17;114(26):5279-89. doi: 10.1182/blood-2009-05-222307. Epub 2009 Oct 26. Blood. 2009. PMID: 19858498
-
Hematopoietic stem cell emergence in the conceptus and the role of Runx1.Int J Dev Biol. 2010;54(6-7):1151-63. doi: 10.1387/ijdb.103106gs. Int J Dev Biol. 2010. PMID: 20711992 Free PMC article. Review.
-
AML1/Runx1 as a versatile regulator of hematopoiesis: regulation of its function and a role in adult hematopoiesis.Int J Hematol. 2006 Aug;84(2):136-42. doi: 10.1532/IJH97.06070. Int J Hematol. 2006. PMID: 16926135 Review.
Cited by
-
Loss of RUNX1/AML1 arginine-methylation impairs peripheral T cell homeostasis.Br J Haematol. 2015 Sep;170(6):859-73. doi: 10.1111/bjh.13499. Epub 2015 May 26. Br J Haematol. 2015. PMID: 26010396 Free PMC article.
-
Intracellular Signaling Pathways Involved in Childhood Acute Lymphoblastic Leukemia; Molecular Targets.Indian J Hematol Blood Transfus. 2016 Jun;32(2):141-53. doi: 10.1007/s12288-015-0609-z. Epub 2015 Oct 20. Indian J Hematol Blood Transfus. 2016. PMID: 27065575 Free PMC article. Review.
-
An AML1-ETO/miR-29b-1 regulatory circuit modulates phenotypic properties of acute myeloid leukemia cells.Oncotarget. 2017 Jun 20;8(25):39994-40005. doi: 10.18632/oncotarget.18127. Oncotarget. 2017. PMID: 28611288 Free PMC article.
-
Inhibition of the RUNX1-CBFβ transcription factor complex compromises mammary epithelial cell identity: a phenotype potentially stabilized by mitotic gene bookmarking.Oncotarget. 2020 Jun 30;11(26):2512-2530. doi: 10.18632/oncotarget.27637. eCollection 2020 Jun 30. Oncotarget. 2020. PMID: 32655837 Free PMC article.
-
Runx1 deficiency permits granulocyte lineage commitment but impairs subsequent maturation.Oncogenesis. 2013 Nov 4;2(11):e78. doi: 10.1038/oncsis.2013.41. Oncogenesis. 2013. PMID: 24189977 Free PMC article.
References
-
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. Prepublished on 1996/01/26 as DOI. - PubMed
-
- North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22(2):158–168. Prepublished on 2004/03/03 as DOI 10.1634/stemcells.22-2-158. - PubMed
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059–1064. Prepublished on 1997/11/14 as DOI. - PubMed
-
- Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103(6):2316–2324. Prepublished on 2003/11/15 as DOI 10.1182/blood-2003-09-3074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

