The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text
- PMID: 23823062
- PMCID: PMC3688812
- DOI: 10.1371/journal.pone.0065390
The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text
Abstract
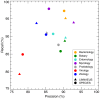
The exponential growth of the biomedical literature is making the need for efficient, accurate text-mining tools increasingly clear. The identification of named biological entities in text is a central and difficult task. We have developed an efficient algorithm and implementation of a dictionary-based approach to named entity recognition, which we here use to identify names of species and other taxa in text. The tool, SPECIES, is more than an order of magnitude faster and as accurate as existing tools. The precision and recall was assessed both on an existing gold-standard corpus and on a new corpus of 800 abstracts, which were manually annotated after the development of the tool. The corpus comprises abstracts from journals selected to represent many taxonomic groups, which gives insights into which types of organism names are hard to detect and which are easy. Finally, we have tagged organism names in the entire Medline database and developed a web resource, ORGANISMS, that makes the results accessible to the broad community of biologists. The SPECIES software is open source and can be downloaded from http://species.jensenlab.org along with dictionary files and the manually annotated gold-standard corpus. The ORGANISMS web resource can be found at http://organisms.jensenlab.org.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

