Responses of soil bacterial and fungal communities to extreme desiccation and rewetting
- PMID: 23823489
- PMCID: PMC3806258
- DOI: 10.1038/ismej.2013.104
Responses of soil bacterial and fungal communities to extreme desiccation and rewetting
Abstract
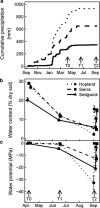
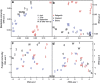
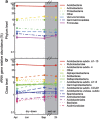
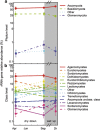
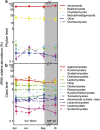
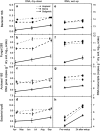
The microbial response to summer desiccation reflects adaptation strategies, setting the stage for a large rainfall-induced soil CO2 pulse upon rewetting, an important component of the ecosystem carbon budget. In three California annual grasslands, the present (DNA-based) and potentially active (RNA-based) soil bacterial and fungal communities were tracked over a summer season and in response to controlled rewetting of intact soil cores. Phylogenetic marker genes for bacterial (16S) and fungal (28S) RNA and DNA were sequenced, and the abundances of these genes and transcripts were measured. Although bacterial community composition differed among sites, all sites shared a similar response pattern of the present and potentially active bacterial community to dry-down and wet-up. In contrast, the fungal community was not detectably different among sites, and was largely unaffected by dry-down, showing marked resistance to dessication. The potentially active bacterial community changed significantly as summer dry-down progressed, then returned to pre-dry-down composition within several hours of rewetting, displaying spectacular resilience. Upon rewetting, transcript copies of bacterial rpoB genes increased consistently, reflecting rapid activity resumption. Acidobacteria and Actinobacteria were the most abundant phyla present and potentially active, and showed the largest changes in relative abundance. The relative increase (Actinobacteria) and decrease (Acidobacteria) with dry-down, and the reverse responses to rewetting reflected a differential response, which was conserved at the phylum level and consistent across sites. These contrasting desiccation-related bacterial life-strategies suggest that predicted changes in precipitation patterns may affect soil nutrient and carbon cycling by differentially impacting activity patterns of microbial communities.
Figures

Similar articles
-
Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate.ISME J. 2015 Mar 17;9(4):946-57. doi: 10.1038/ismej.2014.192. ISME J. 2015. PMID: 25314319 Free PMC article.
-
Resistance and resilience of benthic biofilm communities from a temperate saltmarsh to desiccation and rewetting.ISME J. 2011 Jan;5(1):30-41. doi: 10.1038/ismej.2010.91. Epub 2010 Jul 1. ISME J. 2011. PMID: 20596071 Free PMC article.
-
A Drying-Rewetting Cycle Imposes More Important Shifts on Soil Microbial Communities than Does Reduced Precipitation.mSystems. 2022 Aug 30;7(4):e0024722. doi: 10.1128/msystems.00247-22. Epub 2022 Jun 28. mSystems. 2022. PMID: 35762785 Free PMC article.
-
Soil microbial community responses to multiple experimental climate change drivers.Appl Environ Microbiol. 2010 Feb;76(4):999-1007. doi: 10.1128/AEM.02874-09. Epub 2009 Dec 18. Appl Environ Microbiol. 2010. PMID: 20023089 Free PMC article.
-
Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil.Ecology. 2014 May;95(5):1162-72. doi: 10.1890/13-1031.1. Ecology. 2014. PMID: 25000748
Cited by
-
Soil Microbial Community and Its Interaction with Soil Carbon Dynamics Following a Wetland Drying Process in Mu Us Sandy Land.Int J Environ Res Public Health. 2020 Jun 12;17(12):4199. doi: 10.3390/ijerph17124199. Int J Environ Res Public Health. 2020. PMID: 32545542 Free PMC article.
-
Drought and global hunger: biotechnological interventions in sustainability and management.Planta. 2022 Oct 11;256(5):97. doi: 10.1007/s00425-022-04006-x. Planta. 2022. PMID: 36219256 Review.
-
Microbial response to simulated global change is phylogenetically conserved and linked with functional potential.ISME J. 2016 Jan;10(1):109-18. doi: 10.1038/ismej.2015.96. Epub 2015 Jun 5. ISME J. 2016. PMID: 26046258 Free PMC article.
-
Seasonal Changes in the Soil Microbial Community Structure in Urban Forests.Biology (Basel). 2024 Jan 5;13(1):31. doi: 10.3390/biology13010031. Biology (Basel). 2024. PMID: 38248462 Free PMC article.
-
Volatile and Dissolved Organic Carbon Sources Have Distinct Effects on Microbial Activity, Nitrogen Content, and Bacterial Communities in Soil.Microb Ecol. 2023 Feb;85(2):659-668. doi: 10.1007/s00248-022-01967-0. Epub 2022 Jan 31. Microb Ecol. 2023. PMID: 35102425
References
-
- Alvarez HM, Silva RA, Cesari AC, Zamit AL, Peressutti SR, Reichelt R, et al. Physiological and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol Ecol. 2004;50:75–86. - PubMed
-
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
-
- Bates ST, Garcia-Pichel F. A culture-independent study of free-living fungi in biological soil crusts of the Colorado Plateau: their diversity and relative contribution to microbial biomass. Environ Microbiol. 2009;11:56–67. - PubMed
-
- Birch HF. The effect of soil drying on humus decomposition and nitrogen availability. Plant Soil. 1958;10:9–31.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

