Transgenic Tobacco Overexpressing Tea cDNA Encoding Dihydroflavonol 4-Reductase and Anthocyanidin Reductase Induces Early Flowering and Provides Biotic Stress Tolerance
- PMID: 23823500
- PMCID: PMC3688816
- DOI: 10.1371/journal.pone.0065535
Transgenic Tobacco Overexpressing Tea cDNA Encoding Dihydroflavonol 4-Reductase and Anthocyanidin Reductase Induces Early Flowering and Provides Biotic Stress Tolerance
Abstract
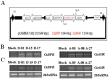
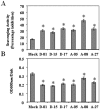
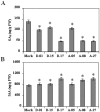
Flavan-3-ols contribute significantly to flavonoid content of tea (Camellia sinensis L.). Dihydroflavonol 4-reductase (DFR) and anthocyanidin reductase (ANR) are known to be key regulatory enzymes of flavan-3-ols biosynthesis. In this study, we have generated the transgenic tobacco overexpressing individually tea cDNA CsDFR and CsANR encoding for DFR and ANR to evaluate their influence on developmental and protective abilities of plant against biotic stress. The transgenic lines of CsDFR and CsANR produced early flowering and better seed yield. Both types of transgenic tobacco showed higher content of flavonoids than control. Flavan-3-ols such as catechin, epicatechin and epicatechingallate were found to be increased in transgenic lines. The free radical scavenging activity of CsDFR and CsANR transgenic lines was improved. Oxidative stress was observed to induce lesser cell death in transgenic lines compared to control tobacco plants. Transgenic tobacco overexpressing CsDFR and CsANR also showed resistance against infestation by a tobacco leaf cutworm Spodoptera litura. Results suggested that the overexpression of CsDFR and CsANR cDNA in tobacco has improved flavonoids content and antioxidant potential. These attributes in transgenic tobacco have ultimately improved their growth and development, and biotic stress tolerance.
Conflict of interest statement
Figures




Similar articles
-
Pyramiding of tea Dihydroflavonol reductase and Anthocyanidin reductase increases flavan-3-ols and improves protective ability under stress conditions in tobacco.3 Biotech. 2017 Jul;7(3):177. doi: 10.1007/s13205-017-0819-1. Epub 2017 Jun 29. 3 Biotech. 2017. PMID: 28664364 Free PMC article.
-
Overexpression of CsANR increased flavan-3-ols and decreased anthocyanins in transgenic tobacco.Mol Biotechnol. 2013 Jun;54(2):426-35. doi: 10.1007/s12033-012-9580-1. Mol Biotechnol. 2013. PMID: 22872496
-
Metabolic Characterization of the Anthocyanidin Reductase Pathway Involved in the Biosynthesis of Flavan-3-ols in Elite Shuchazao Tea (Camellia sinensis) Cultivar in the Field.Molecules. 2017 Dec 15;22(12):2241. doi: 10.3390/molecules22122241. Molecules. 2017. PMID: 29244739 Free PMC article.
-
Increase in flavan-3-ols by silencing flavonol synthase mRNA affects the transcript expression and activity levels of antioxidant enzymes in tobacco.Plant Biol (Stuttg). 2012 Sep;14(5):725-33. doi: 10.1111/j.1438-8677.2011.00550.x. Epub 2012 Feb 10. Plant Biol (Stuttg). 2012. PMID: 22324650
-
Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco.Plant Mol Biol. 2014 Aug;85(6):551-73. doi: 10.1007/s11103-014-0203-z. Epub 2014 Jun 1. Plant Mol Biol. 2014. PMID: 24880475
Cited by
-
Advances in Biosynthesis and Biological Functions of Proanthocyanidins in Horticultural Plants.Foods. 2020 Nov 30;9(12):1774. doi: 10.3390/foods9121774. Foods. 2020. PMID: 33265960 Free PMC article. Review.
-
Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq.Funct Integr Genomics. 2016 Jul;16(4):383-98. doi: 10.1007/s10142-016-0491-2. Epub 2016 Apr 20. Funct Integr Genomics. 2016. PMID: 27098524
-
Comprehensive analysis of putative dihydroflavonol 4-reductase gene family in tea plant.PLoS One. 2019 Dec 26;14(12):e0227225. doi: 10.1371/journal.pone.0227225. eCollection 2019. PLoS One. 2019. PMID: 31877197 Free PMC article.
-
Dissection of Chemical Composition and Associated Gene Expression in the Pigment-Deficient Tea Cultivar 'Xiaoxueya' Reveals an Albino Phenotype and Metabolite Formation.Front Plant Sci. 2019 Nov 27;10:1543. doi: 10.3389/fpls.2019.01543. eCollection 2019. Front Plant Sci. 2019. PMID: 31827483 Free PMC article.
-
Functional Analysis of Genes GlaDFR1 and GlaDFR2 Encoding Dihydroflavonol 4-Reductase (DFR) in Gentiana lutea L. Var. Aurantiaca (M. Laínz) M. Laínz.Biomed Res Int. 2022 Jan 10;2022:1382604. doi: 10.1155/2022/1382604. eCollection 2022. Biomed Res Int. 2022. PMID: 35047628 Free PMC article.
References
-
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, et al. (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483. - PubMed
-
- Tian Li, Pang Y, Dixon R (2008) Biosynthesis and genetic engineering of proanthocyanidins and (iso)flavonoids. Phytochem Rev 7: 445–465.
-
- Dixon RA, Xie D, Sharma SB (2006) Proanthocyanidins-a final frontier in flavonoid research? New Phytol 165: 9–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

