The Observation of Highly Ordered Domains in Membranes with Cholesterol
- PMID: 23823623
- PMCID: PMC3688844
- DOI: 10.1371/journal.pone.0066162
The Observation of Highly Ordered Domains in Membranes with Cholesterol
Abstract
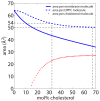
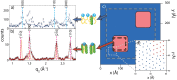
Rafts, or functional domains, are transient nano- or mesoscopic structures in the exoplasmic leaflet of the plasma membrane, and are thought to be essential for many cellular processes. Using neutron diffraction and computer modelling, we present evidence for the existence of highly ordered lipid domains in the cholesterol-rich (32.5 mol%) liquid-ordered ([Formula: see text]) phase of dipalmitoylphosphatidylcholine membranes. The liquid ordered phase in one-component lipid membranes has previously been thought to be a homogeneous phase. The presence of highly ordered lipid domains embedded in a disordered lipid matrix implies non-uniform distribution of cholesterol between the two phases. The experimental results are in excellent agreement with recent computer simulations of DPPC/cholesterol complexes [Meinhardt, Vink and Schmid (2013). Proc Natl Acad Sci USA 110(12): 4476-4481], which reported the existence of nanometer size [Formula: see text] domains in a liquid disordered lipid environment.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

