USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing
- PMID: 23824326
- PMCID: PMC3731547
- DOI: 10.1101/gad.211037.112
USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing
Abstract
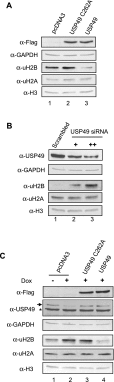
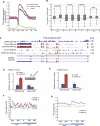
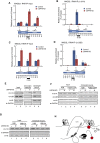
Post-translational histone modifications play important roles in regulating chromatin structure and function. Histone H2B ubiquitination and deubiquitination have been implicated in transcriptional regulation, but the function of H2B deubiquitination is not well defined, particularly in higher eukaryotes. Here we report the purification of ubiquitin-specific peptidase 49 (USP49) as a histone H2B-specific deubiquitinase and demonstrate that H2B deubiquitination by USP49 is required for efficient cotranscriptional splicing of a large set of exons. USP49 forms a complex with RuvB-like1 (RVB1) and SUG1 and specifically deubiquitinates histone H2B in vitro and in vivo. USP49 knockdown results in small changes in gene expression but affects the abundance of >9000 isoforms. Exons down-regulated in USP49 knockdown cells show both elevated levels of alternative splicing and a general decrease in splicing efficiency. Importantly, USP49 is relatively enriched at this set of exons. USP49 knockdown increased H2B ubiquitination (uH2B) levels at these exons as well as upstream 3' and downstream 5' intronic splicing elements. Change in H2B ubiquitination level, as modulated by USP49, regulates U1A and U2B association with chromatin and binding to nascent pre-mRNA. Although H3 levels are relatively stable after USP49 depletion, H2B levels at these exons are dramatically increased, suggesting that uH2B may enhance nucleosome stability. Therefore, this study identifies USP49 as a histone H2B-specific deubiquitinase and uncovers a critical role for H2B deubiquitination in cotranscriptional pre-mRNA processing events.
Keywords: H2B deubiquitination; USP49; pre-mRNA splicing; transcription.
Figures

References
-
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 - PubMed
-
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L 2011. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
