Non-M variants of human immunodeficiency virus type 1
- PMID: 23824367
- PMCID: PMC3719493
- DOI: 10.1128/CMR.00012-13
Non-M variants of human immunodeficiency virus type 1
Abstract
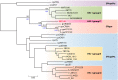
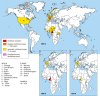
The AIDS pandemic that started in the early 1980s is due to human immunodeficiency virus type 1 (HIV-1) group M (HIV-M), but apart from this major group, many divergent variants have been described (HIV-1 groups N, O, and P and HIV-2). The four HIV-1 groups arose from independent cross-species transmission of the simian immunodeficiency viruses (SIVs) SIVcpz, infecting chimpanzees, and SIVgor, infecting gorillas. This, together with human adaptation, accounts for their genomic, phylogenetic, and virological specificities. Nevertheless, the natural course of non-M HIV infection seems similar to that of HIV-M. The virological monitoring of infected patients is now possible with commercial kits, but their therapeutic management remains complex. All non-M variants were principally described for patients linked to Cameroon, where HIV-O accounts for 1% of all HIV infections; only 15 cases of HIV-N infection and 2 HIV-P infections have been reported. Despite improvements in our knowledge, many fascinating questions remain concerning the origin, genetic evolution, and slow spread of these variants. Other variants may already exist or may arise in the future, calling for close surveillance. This review provides a comprehensive, up-to-date summary of the current knowledge on these pathogens, including the historical background of their discovery; the latest advances in the comprehension of their origin and spread; and clinical, therapeutic, and laboratory aspects that may be useful for the management and the treatment of patients infected with these divergent viruses.
Figures








References
-
- Barré-Sinoussi F, Chermann JC, Rey P, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Brun-Vezinet F, Rouzioux C, Rozenbaum W, Montagnier L. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871 - PubMed
-
- Barin F, M'Boup S, Denis F, Kanki P, Allan JS, Lee TH, Essex M. 1985. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of West Africa. Lancet ii:1387–1389 - PubMed
-
- Clavel F, Guétard D, Brun-Vézinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, Klatzmann D, Champalimaud JL, Montagnier L. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343–346 - PubMed
-
- De Leys R, Vanderborght B, Vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems B, Taelman H, van der Groen G, Piot P, Tersmette T, Huisman JG, van Heuverswyn H. 1990. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of West-Central African origin. J. Virol. 64:1207–1216 - PMC - PubMed
-
- Nkengasong JN, Peeters M, vanden Haesevelde M, Musi SS, Willems B, Ndumbe PM, Delaporte E, Perret JL, Piot P, van den Groen G. 1993. Antigenic evidence of the presence of the aberrant HIV-1ant70 virus in Cameroon and Gabon. AIDS 7:1536–1538 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

