Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology
- PMID: 23824373
- PMCID: PMC3719498
- DOI: 10.1128/CMR.00072-12
Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology
Abstract
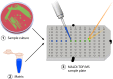
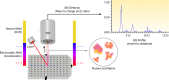
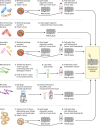
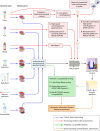
Within the past decade, clinical microbiology laboratories experienced revolutionary changes in the way in which microorganisms are identified, moving away from slow, traditional microbial identification algorithms toward rapid molecular methods and mass spectrometry (MS). Historically, MS was clinically utilized as a high-complexity method adapted for protein-centered analysis of samples in chemistry and hematology laboratories. Today, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS is adapted for use in microbiology laboratories, where it serves as a paradigm-shifting, rapid, and robust method for accurate microbial identification. Multiple instrument platforms, marketed by well-established manufacturers, are beginning to displace automated phenotypic identification instruments and in some cases genetic sequence-based identification practices. This review summarizes the current position of MALDI-TOF MS in clinical research and in diagnostic clinical microbiology laboratories and serves as a primer to examine the "nuts and bolts" of MALDI-TOF MS, highlighting research associated with sample preparation, spectral analysis, and accuracy. Currently available MALDI-TOF MS hardware and software platforms that support the use of MALDI-TOF with direct and precultured specimens and integration of the technology into the laboratory workflow are also discussed. Finally, this review closes with a prospective view of the future of MALDI-TOF MS in the clinical microbiology laboratory to accelerate diagnosis and microbial identification to improve patient care.
Figures




References
-
- Wassenberg MWM, Kluytmans JAJW, Box ATA, Bosboom RW, Buiting AGM, Van Elzakker EPM, Melchers WJG, Van Rijen MML, Thijsen SFT, Troelstra A, Vandenbroucke-Grauls CMJE, Visser CE, Voss A, Wolffs PFG, Wulf MWH, Van Zwet AA, De Wit GA, Bonten MJM. 2010. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin. Microbiol. Infect. 16:1754–1761 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

