Influenza A virus hemagglutinin trimerization completes monomer folding and antigenicity
- PMID: 23824811
- PMCID: PMC3754138
- DOI: 10.1128/JVI.00471-13
Influenza A virus hemagglutinin trimerization completes monomer folding and antigenicity
Abstract
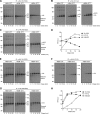
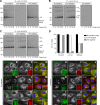
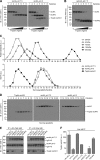
Influenza A virus (IAV) remains an important human pathogen largely because of antigenic drift, the rapid emergence of antibody escape mutants that precludes durable vaccination. The most potent neutralizing antibodies interact with cognate epitopes in the globular "head" domain of hemagglutinin (HA), a homotrimeric glycoprotein. The H1 HA possesses five distinct regions defined by a large number of mouse monoclonal antibodies (MAbs), i.e., Ca1, Ca2, Cb, Sa, and Sb. Ca1-Ca2 sites require HA trimerization to attain full antigenicity, consistent with their locations on opposite sides of the trimer interface. Here, we show that full antigenicity of Cb and Sa sites also requires HA trimerization, as revealed by immunofluorescence microscopy of IAV-infected cells and biochemically by pulse-chase radiolabeling experiments. Surprisingly, epitope antigenicity acquired by HA trimerization persists following acid triggering of the globular domains dissociation and even after proteolytic release of monomeric heads from acid-treated HA. Thus, the requirement for HA trimerization by trimer-specific MAbs mapping to the Ca, Cb, and Sa sites is not dependent upon the bridging of adjacent monomers in the native HA trimer. Rather, complete antigenicity of HA (and, by inference, immunogenicity) requires a final folding step that accompanies its trimerization. Once this conformational change occurs, HA trimers themselves would not necessarily be required to induce a highly diverse neutralizing response to epitopes in the globular domain.
Figures


Similar articles
-
Biogenesis of influenza a virus hemagglutinin cross-protective stem epitopes.PLoS Pathog. 2014 Jun 12;10(6):e1004204. doi: 10.1371/journal.ppat.1004204. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24945804 Free PMC article.
-
Epitope mapping of the 2009 pandemic and the A/Brisbane/59/2007 seasonal (H1N1) influenza virus haemagglutinins using mAbs and escape mutants.J Gen Virol. 2014 Nov;95(Pt 11):2377-2389. doi: 10.1099/vir.0.067819-0. Epub 2014 Jul 30. J Gen Virol. 2014. PMID: 25078301
-
Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies.J Virol. 2007 Dec;81(23):12911-7. doi: 10.1128/JVI.01522-07. Epub 2007 Sep 19. J Virol. 2007. PMID: 17881439 Free PMC article.
-
N-linked glycosylation in the hemagglutinin of influenza A viruses.Yonsei Med J. 2012 Sep;53(5):886-93. doi: 10.3349/ymj.2012.53.5.886. Yonsei Med J. 2012. PMID: 22869469 Free PMC article. Review.
-
The antigenic architecture of the hemagglutinin of influenza H5N1 viruses.Mol Immunol. 2013 Dec;56(4):705-19. doi: 10.1016/j.molimm.2013.07.010. Epub 2013 Aug 7. Mol Immunol. 2013. PMID: 23933511 Review.
Cited by
-
Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity.Elife. 2015 Aug 7;4:e07467. doi: 10.7554/eLife.07467. Elife. 2015. PMID: 26252514 Free PMC article.
-
Generation of a protective murine monoclonal antibody against the stem of influenza hemagglutinins from group 1 viruses and identification of resistance mutations against it.PLoS One. 2019 Sep 12;14(9):e0222436. doi: 10.1371/journal.pone.0222436. eCollection 2019. PLoS One. 2019. PMID: 31513662 Free PMC article.
-
The Encapsulation of Hemagglutinin in Protein Bodies Achieves a Stronger Immune Response in Mice than the Soluble Antigen.Front Plant Sci. 2016 Feb 16;7:142. doi: 10.3389/fpls.2016.00142. eCollection 2016. Front Plant Sci. 2016. PMID: 26909090 Free PMC article.
-
A Recombinant Mosaic HAs Influenza Vaccine Elicits Broad-Spectrum Immune Response and Protection of Influenza a Viruses.Vaccines (Basel). 2024 Sep 2;12(9):1008. doi: 10.3390/vaccines12091008. Vaccines (Basel). 2024. PMID: 39340038 Free PMC article.
-
Effect of Antigen Structure in Subunit Vaccine Nanoparticles on Humoral Immune Responses.ACS Biomater Sci Eng. 2023 Mar 13;9(3):1296-1306. doi: 10.1021/acsbiomaterials.2c01516. Epub 2023 Feb 27. ACS Biomater Sci Eng. 2023. PMID: 36848229 Free PMC article.
References
-
- Hers JF. 1966. Disturbances of the ciliated epithelium due to influenza virus. Am. Rev. Respir. Dis. 93(Suppl):162–177 - PubMed
-
- Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 - PubMed
-
- Copeland CS, Zimmer KP, Wagner KR, Healey GA, Mellman I, Helenius A. 1988. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell 53:197–209 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

