p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1
- PMID: 23825024
- PMCID: PMC3756925
- DOI: 10.1091/mbc.E12-10-0771
p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1
Abstract
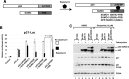
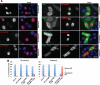
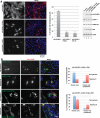
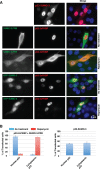
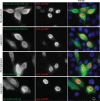
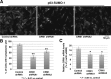
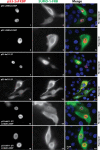
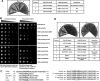
Chromosomal region maintenance 1 (CRM1) mediates p53 nuclear export. Although p53 SUMOylation promotes its nuclear export, the underlying mechanism is unclear. Here we show that tethering of a small, ubiquitin-like modifier (SUMO) moiety to p53 markedly increases its cytoplasmic localization. SUMO attachment to p53 does not affect its oligomerization, suggesting that subunit dissociation required for exposing p53's nuclear export signal (NES) is unnecessary for p53 nuclear export. Surprisingly, SUMO-mediated p53 nuclear export depends on the SUMO-interacting motif (SIM)-binding pocket of SUMO-1. The CRM1 C-terminal domain lacking the NES-binding groove interacts with tetrameric p53, and the proper folding of the p53 core domain, rather than the presence of the N- or C-terminal tails, appears to be important for p53-CRM1 interaction. The CRM1 Huntington, EF3, a subunit of PP2A, and TOR1 9 (HEAT9) loop, which regulates GTP-binding nuclear protein Ran binding and cargo release, contains a prototypical SIM. Remarkably, disruption of this SIM in conjunction with a mutated SIM-binding groove of SUMO-1 markedly enhances the binding of CRM1 to p53-SUMO-1 and their accumulation in the nuclear pore complexes (NPCs), as well as their persistent association in the cytoplasm. We propose that SUMOylation of a CRM1 cargo such as p53 at the NPCs unlocks the HEAT9 loop of CRM1 to facilitate the disassembly of the transporting complex and cargo release to the cytoplasm.
Figures



Similar articles
-
The Cellular Distribution of RanGAP1 Is Regulated by CRM1-Mediated Nuclear Export in Mammalian Cells.PLoS One. 2015 Oct 27;10(10):e0141309. doi: 10.1371/journal.pone.0141309. eCollection 2015. PLoS One. 2015. PMID: 26506250 Free PMC article.
-
An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1.EMBO J. 2010 Jun 16;29(12):2002-13. doi: 10.1038/emboj.2010.89. Epub 2010 May 18. EMBO J. 2010. PMID: 20485264 Free PMC article.
-
A 2.1-Å-resolution crystal structure of unliganded CRM1 reveals the mechanism of autoinhibition.J Mol Biol. 2013 Jan 23;425(2):350-64. doi: 10.1016/j.jmb.2012.11.014. Epub 2012 Nov 16. J Mol Biol. 2013. PMID: 23164569
-
Atomic basis of CRM1-cargo recognition, release and inhibition.Semin Cancer Biol. 2014 Aug;27:52-61. doi: 10.1016/j.semcancer.2014.03.002. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631835 Free PMC article. Review.
-
Inhibition of CRM1-dependent nuclear export sensitizes malignant cells to cytotoxic and targeted agents.Semin Cancer Biol. 2014 Aug;27:62-73. doi: 10.1016/j.semcancer.2014.03.001. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631834 Free PMC article. Review.
Cited by
-
The Four Homeostasis Knights: In Balance upon Post-Translational Modifications.Int J Mol Sci. 2022 Nov 21;23(22):14480. doi: 10.3390/ijms232214480. Int J Mol Sci. 2022. PMID: 36430960 Free PMC article. Review.
-
CRISPR-Cas9 Screening of Kaposi's Sarcoma-Associated Herpesvirus-Transformed Cells Identifies XPO1 as a Vulnerable Target of Cancer Cells.mBio. 2019 May 14;10(3):e00866-19. doi: 10.1128/mBio.00866-19. mBio. 2019. PMID: 31088931 Free PMC article.
-
Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway.Nat Commun. 2018 Jan 10;9(1):143. doi: 10.1038/s41467-017-02413-3. Nat Commun. 2018. PMID: 29321472 Free PMC article.
-
Dietary flavonoid fisetin binds human SUMO1 and blocks sumoylation of p53.PLoS One. 2020 Jun 12;15(6):e0234468. doi: 10.1371/journal.pone.0234468. eCollection 2020. PLoS One. 2020. PMID: 32530958 Free PMC article.
-
Cell fate regulation governed by p53: Friends or reversible foes in cancer therapy.Cancer Commun (Lond). 2024 Mar;44(3):297-360. doi: 10.1002/cac2.12520. Epub 2024 Feb 4. Cancer Commun (Lond). 2024. PMID: 38311377 Free PMC article. Review.
References
-
- Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. - PubMed
-
- Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. - PubMed
-
- Carter S, Vousden KH. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle. 2008;7:2519–2528. - PubMed
-
- Chang CC, et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell. 2011;42:62–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

