Genomic signatures predict poor outcome in undifferentiated pleomorphic sarcomas and leiomyosarcomas
- PMID: 23825676
- PMCID: PMC3692486
- DOI: 10.1371/journal.pone.0067643
Genomic signatures predict poor outcome in undifferentiated pleomorphic sarcomas and leiomyosarcomas
Abstract
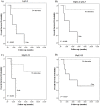
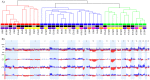
Undifferentiated high-grade pleomorphic sarcomas (UPSs) display aggressive clinical behavior and frequently develop local recurrence and distant metastasis. Because these sarcomas often share similar morphological patterns with other tumors, particularly leiomyosarcomas (LMSs), classification by exclusion is frequently used. In this study, array-based comparative genomic hybridization (array CGH) was used to analyze 20 UPS and 17 LMS samples from untreated patients. The LMS samples presented a lower frequency of genomic alterations compared with the UPS samples. The most frequently altered UPS regions involved gains at 20q13.33 and 7q22.1 and losses at 3p26.3. Gains at 8q24.3 and 19q13.12 and losses at 9p21.3 were frequently detected in the LMS samples. Of these regions, gains at 1q21.3, 11q12.2-q12.3, 16p11.2, and 19q13.12 were significantly associated with reduced overall survival times in LMS patients. A multivariate analysis revealed that gains at 1q21.3 were an independent prognostic marker of shorter survival times in LMS patients (HR = 13.76; P = 0.019). Although the copy number profiles of the UPS and LMS samples could not be distinguished using unsupervised hierarchical clustering analysis, one of the three clusters presented cases associated with poor prognostic outcome (P = 0.022). A relative copy number analysis for the ARNT, SLC27A3, and PBXIP1 genes was performed using quantitative real-time PCR in 11 LMS and 16 UPS samples. Gains at 1q21-q22 were observed in both tumor types, particularly in the UPS samples. These findings provide strong evidence for the existence of a genomic signature to predict poor outcome in a subset of UPS and LMS patients.
Conflict of interest statement
Figures

References
-
- Osuna D, de Alava E (2009) Molecular pathology of sarcomas. Rev Recent Clin Trials 4: 12–26. - PubMed
-
- Wardelmann E, Schildhaus HU, Merkelbach-Bruse S, Hartmann W, Reichardt P, et al. (2010) Soft tissue sarcoma: from molecular diagnosis to selection of treatment. Pathological diagnosis of soft tissue sarcoma amid molecular biology and targeted therapies. Ann Oncol 21: 265–269. - PubMed
-
- Fletcher CD (2006) The evolving classification of soft tissue tumours: an update based on the new WHO classification. Histopathology 48: 3–12. - PubMed
-
- Nascimento AF, Raut CP (2008) Diagnosis and management of pleomorphic sarcomas (So-Called “MFH”) in adults. J Surg Oncol 97: 330–339. - PubMed
-
- Erlandson RA, Antonescu CR (2004) The rise and fall of malignant fibrous histiocytoma. Ultrastruct Pathol 28: 283–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

