Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing
- PMID: 23825951
- PMCID: PMC3694860
- DOI: 10.1371/journal.ppat.1003460
Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing
Abstract
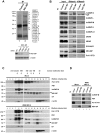
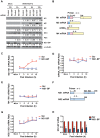
Influenza A virus is a major human pathogen with a genome comprised of eight single-strand, negative-sense, RNA segments. Two viral RNA segments, NS1 and M, undergo alternative splicing and yield several proteins including NS1, NS2, M1 and M2 proteins. However, the mechanisms or players involved in splicing of these viral RNA segments have not been fully studied. Here, by investigating the interacting partners and function of the cellular protein NS1-binding protein (NS1-BP), we revealed novel players in the splicing of the M1 segment. Using a proteomics approach, we identified a complex of RNA binding proteins containing NS1-BP and heterogeneous nuclear ribonucleoproteins (hnRNPs), among which are hnRNPs involved in host pre-mRNA splicing. We found that low levels of NS1-BP specifically impaired proper alternative splicing of the viral M1 mRNA segment to yield the M2 mRNA without affecting splicing of mRNA3, M4, or the NS mRNA segments. Further biochemical analysis by formaldehyde and UV cross-linking demonstrated that NS1-BP did not interact directly with viral M1 mRNA but its interacting partners, hnRNPs A1, K, L, and M, directly bound M1 mRNA. Among these hnRNPs, we identified hnRNP K as a major mediator of M1 mRNA splicing. The M1 mRNA segment generates the matrix protein M1 and the M2 ion channel, which are essential proteins involved in viral trafficking, release into the cytoplasm, and budding. Thus, reduction of NS1-BP and/or hnRNP K levels altered M2/M1 mRNA and protein ratios, decreasing M2 levels and inhibiting virus replication. Thus, NS1-BP-hnRNPK complex is a key mediator of influenza A virus gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- WHO (April 2009) http://www.who.int/mediacentre/factsheets/fs211/en/index.html. Fact sheet Number 211.
-
- Palese P, Shaw ML (2007) Orthomyxoviridae: The virus and their replication. In: Knipe, D.M, et al.. Eds. Fields Virology. 5th edition. Philadelphia, USA: Lippincott Williams & Wilkins. 1647–1689.
-
- Palese P, Shaw ML (2007) Orthomyxoviridae: The virus and their replication. In: Knipe, D.M, et al.. Eds. Fields Virology. 5th edition. Lippincott Williams & Wilkins, Philadelphia, USA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

